Transcriptional control by the TGF-beta/Smad signaling system - PubMed (original) (raw)
Review
Transcriptional control by the TGF-beta/Smad signaling system
J Massagué et al. EMBO J. 2000.
No abstract available
Figures
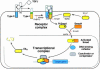
Fig. 1. Schematic representation of the TGF-β/Smad signaling engine. This system involves a family of membrane receptor protein kinases and a family of receptor substrates (the Smad proteins) that march into the nucleus where they act as transcription factors. The ligand TGF-β assembles a receptor complex that activates Smads, and the Smads assemble multisubunit complexes that regulate transcription. Two general steps suffice to carry the hormonal stimulus to target genes. The central components of this signaling system are indicated along with the sites of action of various positive and negative regulators. See the text for further details.
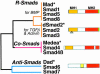
Fig. 2. The Smad family. Simplified dendrogram of sequence similarity between the three Smad subfamilies. The receptor-regulated Smads (R-Smads) and their cooperating Smads (Co-Smads) contain conserved N-terminal (MH1) and C-terminal (MH2) domains separated by a divergent region. Only the MH2 domain is conserved in the inhibitory Smads (Anti-Smads). The green sliver represents the receptor phosphorylation sites at the extreme C-terminus of the R-Smads. The triangle represents the alternatively spliced insert in Smad2. Asterisks denote representative members from Drosophila.
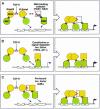
Fig. 3. Smad structural domains and their functions. Representation of the three-dimensional structures of the Smad3 MH1 domain bound to the AGAC sequence, and the Smad2 MH2 domain. The principal interactions of these two domains are listed. The structures involved in these interactions are shown in different colors: the β-hairpin (βhp) that mediated DNA binding, the L3 loop and α-helix 1 (αH-1) that specify Smad interactions with type I receptors, and the α-helix 2 (αH-2) that specifies Smad2 interaction with FAST. SSXS, receptor phosphorylation sites (adapted from Shi et al., 1997, 1998; Wu et al., 2000).
Fig. 4. Making choices through the Smad system.The combinatorial organization of this system as presently understood.
Fig. 5. Smad transcriptional partners. General models for the recognition and regulation of specific target genes by Smads in concert with DNA-binding adaptors such as FAST and OAZ (model A) and constitutive (e.g. TFE and CBF) or signal-regulated (e.g. AP-1) transcription factors that interact with the MH1 domain upon agonist activation (model B) or with the MH2 domain in the basal state (model C). MH1 and MH2, Smad domains; orange boxes, transactivator domains; SID, Smad interaction domain. Although two Smad DNA sites are depicted in each model, only one may be used in certain response elements.
Similar articles
- Transforming growth factor-beta up-regulates the beta 5 integrin subunit expression via Sp1 and Smad signaling.
Lai CF, Feng X, Nishimura R, Teitelbaum SL, Avioli LV, Ross FP, Cheng SL. Lai CF, et al. J Biol Chem. 2000 Nov 17;275(46):36400-6. doi: 10.1074/jbc.M002131200. J Biol Chem. 2000. PMID: 10964912 - Tieg3/Klf11 induces apoptosis in OLI-neu cells and enhances the TGF-beta signaling pathway by transcriptional repression of Smad7.
Gohla G, Krieglstein K, Spittau B. Gohla G, et al. J Cell Biochem. 2008 Jun 1;104(3):850-61. doi: 10.1002/jcb.21669. J Cell Biochem. 2008. PMID: 18189266 - BF-1 interferes with transforming growth factor beta signaling by associating with Smad partners.
Dou C, Lee J, Liu B, Liu F, Massague J, Xuan S, Lai E. Dou C, et al. Mol Cell Biol. 2000 Sep;20(17):6201-11. doi: 10.1128/MCB.20.17.6201-6211.2000. Mol Cell Biol. 2000. PMID: 10938097 Free PMC article. - [Transforming growth factor-beta signaling and cancer].
Miyazono K. Miyazono K. Hum Cell. 2000 Sep;13(3):97-101. Hum Cell. 2000. PMID: 11197777 Review. Japanese. - Signal transduction by the TGF-beta superfamily.
Attisano L, Wrana JL. Attisano L, et al. Science. 2002 May 31;296(5573):1646-7. doi: 10.1126/science.1071809. Science. 2002. PMID: 12040180 Review.
Cited by
- Vascular biology of uterine fibroids: connecting fibroids and vascular disorders.
Kirschen GW, AlAshqar A, Miyashita-Ishiwata M, Reschke L, El Sabeh M, Borahay MA. Kirschen GW, et al. Reproduction. 2021 Jul 8;162(2):R1-R18. doi: 10.1530/REP-21-0087. Reproduction. 2021. PMID: 34034234 Free PMC article. Review. - Signaling pathways in the regulation of cancer stem cells and associated targeted therapy.
Manni W, Min W. Manni W, et al. MedComm (2020). 2022 Oct 5;3(4):e176. doi: 10.1002/mco2.176. eCollection 2022 Dec. MedComm (2020). 2022. PMID: 36226253 Free PMC article. Review. - Gastric tumor development in Smad3-deficient mice initiates from forestomach/glandular transition zone along the lesser curvature.
Nam KT, O'Neal R, Lee YS, Lee YC, Coffey RJ, Goldenring JR. Nam KT, et al. Lab Invest. 2012 Jun;92(6):883-95. doi: 10.1038/labinvest.2012.47. Epub 2012 Mar 12. Lab Invest. 2012. PMID: 22411066 Free PMC article. - Astragaloside IV controls collagen reduction in photoaging skin by improving transforming growth factor-β/Smad signaling suppression and inhibiting matrix metalloproteinase-1.
Chen B, Li R, Yan N, Chen G, Qian W, Jiang HL, Ji C, Bi ZG. Chen B, et al. Mol Med Rep. 2015 May;11(5):3344-8. doi: 10.3892/mmr.2015.3212. Epub 2015 Jan 16. Mol Med Rep. 2015. PMID: 25591734 Free PMC article. - The Regulatory Network of CREB3L1 and Its Roles in Physiological and Pathological Conditions.
Zhao Y, Yu Z, Song Y, Fan L, Lei T, He Y, Hu S. Zhao Y, et al. Int J Med Sci. 2024 Jan 1;21(1):123-136. doi: 10.7150/ijms.90189. eCollection 2024. Int J Med Sci. 2024. PMID: 38164349 Free PMC article. Review.
References
- Akiyoshi S., Inoue,H., Hanai,J., Kusanagi,K., Nemoto,N., Miyazono,K. and Kawabata,M. (1999) c-Ski acts as a transcriptional co-repressor in transforming growth factor-β signaling through interaction with smads. J. Biol. Chem., 274, 35269–35277. - PubMed
- Ashcroft G.S. et al. (1999) Mice lacking Smad3 show accelerated wound healing and an impaired local inflammatory response. Nature Cell Biol., 1, 260–266. - PubMed
- Beckmann H., Su,L.K. and Kadesch,T. (1990) TFE3: a helix–loop–helix protein that activates transcription through the immunoglobulin enhancer muE3 motif. Genes Dev., 4, 167–179. - PubMed
- Bertolino E., Reimund,B., Wildt-Perinic,D. and Clerc,R. (1995) A novel homeobox protein which recognizes a TGT core and functionally interferes with a retinoid-responsive motif. J. Biol. Chem., 270, 31178–31188. - PubMed
- Brown C.B., Boyer,A.S., Runyan,R.B. and Barnett,J.V. (1999) Requirement of type III TGF-β receptor for endocardial cell transformation in the heart. Science, 283, 2080–2082. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases