The p300/CBP acetyltransferases function as transcriptional coactivators of beta-catenin in vertebrates - PubMed (original) (raw)
The p300/CBP acetyltransferases function as transcriptional coactivators of beta-catenin in vertebrates
A Hecht et al. EMBO J. 2000.
Abstract
Wnt growth factors regulate a variety of developmental processes by altering specific gene expression patterns. In vertebrates beta-catenin acts as transcriptional activator, which is needed to overcome target gene repression by Groucho/TLE proteins, and to permit promoter activation as the final consequence of Wnt signaling. However, the molecular mechanisms of transcriptional activation by beta-catenin are only poorly understood. Here we demonstrate that the closely related acetyltransferases p300 and CBP potentiate beta-catenin-mediated activation of the siamois promoter, a known Wnt target. beta-catenin and p300 also synergize to stimulate a synthetic reporter gene construct, whereas activation of the cyclin D1 promoter by beta-catenin is refractory to p300 stimulation. Axis formation and activation of the beta-catenin target genes siamois and Xnr-3 in Xenopus embryos are sensitive to the E1A oncoprotein, a known inhibitor of p300/CBP. The C-terminus of beta-catenin interacts directly with a region overlapping the CH-3 domain of p300. p300 could participate in alleviating promoter repression imposed by chromatin structure and in recruiting the basal transcription machinery to promoters of particular Wnt target genes.
Figures
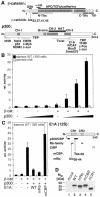
Fig. 1. β-catenin utilizes p300 as coactivator at the siamois promoter. (A) Schematic representation of β-catenin and p300. In β-catenin (hatched bar) the Armadillo repeats (gray boxes) and interaction domains for various binding partners are marked. The GSK-3β target sequence (GSK) and the transactivating elements (N-TAs, C-TAs) are also shown. Autonomous TAs: dark gray; auxiliary TAs: light gray. In β-catS33A serine/threonine residues at positions 33, 37, 41 and 45 in the GSK-3β target sequence were substituted with alanines. In p300 the cysteine/histidine-rich elements CH-1, CH-2, and CH-3, the Bromo domain (gray boxes), the HAT domain (black bar) and regions interacting with nuclear hormone receptors (NHR), other transcription factors and E1A are denoted. (B) 293 cells were transfected with luciferase reporter plasmids harboring either the WT siamois promoter or a deletion mutant lacking the TCF binding sites (1.0 µg), the lacZ expression plasmid pCMVβ (Promega) (0.5 µg), a construct for expression of β-catS33A (0.5 µg) and increasing amounts of expression vector for p300 (0.5, 1.0, 2.0 µg) as indicated. Luciferase activities were determined 40 h post transfection, normalized against β-galactosidase values and expressed as relative activity compared with cells transfected with the WT siamois reporter alone (rel. activity: 1). (C) 293 cells were transfected with the WT siamois reporter, pCMVβ and expression vectors for β-catS33A (0.5 µg), p300 (2.0 µg) and WT E1A or mutant E1A (0.2 µg) as indicated. Reporter activity without β-catenin and p300: 1. A diagram of 12S-E1A is depicted. Conserved regions 1 and 2 (CR1, CR2), domains involved in binding Rb-family members, p300/CBP and the CtBP transcriptional repressor, and the location of the mRb and mCBP mutations are indicated. (B, C) Average values from at least three independent experiments are shown with standard deviations. (D) SW480 cells were transfected with expression vectors (2.5 µg each) for the E1A proteins as indicated. Whole cell lysates were prepared 40 h after transfection and E1A expression levels were analyzed by Western blotting with a monoclonal antibody. Mw: molecular weight markers.
Fig. 2. Promoter-specific differences in transcriptional activation mediated by β-catenin and p300. (A) 293 cells were transfected with the TOPFLASH or FOPFLASH reporter plasmids (1.0 µg), pCMVβ (0.5 µg), a construct for expression of β-catS33A (0.5 µg) and increasing amounts of expression vector for p300 (0.5, 1.0, 2.0 µg) as indicated. Luciferase activities were determined as before. (B) Same experiment as in (A) but with a reporter construct harboring the cyclin D1 promoter. Reporter activity in (A) and (B) without β-catenin and p300: 1.
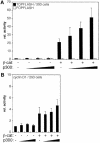
Fig. 3. (A) Endogenous β-catenin in SW480 cells utilizes p300 as coactivator. SW480 cells were transfected with pCMVβ (0.5 µg), the TOPFLASH/FOPFLASH (1.5 µg) or siamois promoter constructs (2.5 µg) and expression vectors for p300 (2.0 µg) or dominant negative ΔN-TCF4 (2.0 µg) as indicated. To interfere with p300-mediated transcriptional activation WT E1A, E1AmRb, E1AmCBP or E1AΔCR1 were cotransfected (0.2 µg each). Luciferase activities were determined as before. Activity of the TOPFLASH or WT siamois reporter without exogenous p300, ΔN-TCF4 or E1A was set as 100% (n.d.: not done). (B) Functional similarity of p300 and CBP. SW480 cells were transfected with pCMVβ (0.5 µg), the TOPFLASH/FOPFLASH reporter constructs (1.5 µg) and expression vectors for p300 (4.0 µg) or CBP (1.0, 2.0 and 4.0 µg) as indicated. Luciferase activities, determined as before, are expressed as relative activities compared with cells transfected with FOPFLASH alone. (C) HAT activity of p300 is not required for its function as coactivator of β-catenin. 293 cells were transfected with the siamois reporter (0.5 µg), pCMVβ (0.1 µg), and expression vectors for β-catS33A (0.5 µg), WT p300, or HAT-defective p300mut (0.5 and 2.5 µg) as indicated. Control transfectants received 2.5 µg of empty expression vector or p300 plasmids. Luciferase activities were determined as before. Reporter activity without β-catenin and p300: 1.
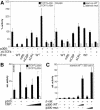
Fig. 4. Interactions between p300 and β-catenin in vitro. (A) Mapping of the β-catenin interaction domain in p300. GST or GST–β-catenin fusion proteins were bound to GSH–Sepharose beads and incubated with [35S]methionine-labeled p300 fragments. After extensive washing, proteins retained by GST or GST–β-catenin were analyzed together with 10% of the input material by SDS–PAGE and fluorography. The structure of p300 and its derivatives is shown schematically. End points of the p300 fragments and the location of the β-catenin interaction domain (hatched bar) are given. (B) Mapping of the p300 interaction domain in β-catenin. Bacterially expressed GST fusion proteins with β-catenin sequences as indicated were bound to GSH–Sepharose and incubated with a radiolabeled p300 fragment (residues 1737–2378). Material bound to the various GST fusions was analyzed by SDS–PAGE and fluorography. Lane 1: a sample corresponding to 10% of the p300 input material. A diagram of β-catenin giving the location of its transactivating elements as well as the interaction sites TI–TIII for TBP (light gray bar), Pontin52 (open bar) and p300 (black bar) is also shown. (C) Direct interactions between β-catenin and p300. GST or GST fusion proteins with p300 sequences as indicated were bound to GSH–Sepharose and incubated with histidine-tagged β-catenin (β-catHis6) purified from E.coli. Material bound to the various GST–p300 fusions was analyzed by SDS–PAGE and Western blotting. (D) The C-terminus of β-catenin but not LEF-1 interacts with the CH-3 domain of p300 in vitro. Bacterially expressed GST or GST fusion proteins with p300 sequences as indicated were bound to GSH–Sepharose and incubated with radiolabeled LEF-1 or a β-catenin fragment comprising residues 553–781. Material retained on the GST or GST–p300-coated matrices was eluted and analyzed by SDS–PAGE and fluorography. Lane 1, Input: a sample corresponding to 10% of the LEF-1 or β-catenin material used for the assay. Mw: molecular weight markers.
Fig. 5. Interactions between p300 and β-catenin in vivo. (A) Co-immunoprecipitation of p300 and β-catenin. 293 cells were transfected with expression plasmids for HA-tagged p300 and myc-tagged β-catS33A as indicated. Cell extracts were prepared 40 h after transfection and used for immunoprecipitations with anti-HA or anti-myc antibodies. Immunoprecipitated material and a fraction of each lysate was resolved by SDS–PAGE and analyzed by Western blotting with antibodies as shown. IP: immunoprecipitation; IB: immunoblot. (B) Mammalian two-hybrid analyses of interactions between β-catenin and p300. NIH 3T3 cells were transfected with pCMVβ (0.05 µg), the Gal4-dependent pG5E1bLuc reporter construct (0.1 µg) and empty control vectors or expression vectors for Gal4–p300 (1737–1891) (0.5 µg), LEFΔN–VP16 (0.5 µg) and β-catenin–VP16 fusions (0.5 µg) as indicated. Luciferase activities, determined as before, are expressed as relative activity compared with cells transfected with the pG5E1bLuc reporter and empty expression vector (rel. activity: 1). The diagram on the right shows the structure of the β-catenin–VP16 and the LEFΔN–VP16 prey fusions, the Gal4–p300 (1737–1891) bait and the pG5E1bLuc reporter. In β-catenin, the GSK recognition sequence, N-terminal and C-terminal transactivating elements (N-TAs, C-TAs) and the p300 interaction domain (black bar) are marked. LEFΔN–VP16 contains the HMG-box (hatched box) but lacks the β-catenin binding site. The Gal4–p300 fusion harbors the C-terminal portion of the CH-3 domain (dark gray box). VP16: VP16 transactivation domain.
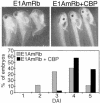
Fig. 6. Formation of the endogenous body axis is sensitive to E1AmRb and can be rescued by coexpression of CBP. Embryos were injected at the four-cell stage in the vegetal zone of the two dorsal blastomeres with RNA for E1AmRb (0.5 ng) alone or together with 2.0 ng of CBP RNA. Ventralization of embryos was scored at the tadpole stage and is expressed as dorso-anterior index (DAI; Kao and Elinson, 1988). Normal embryos have a DAI of 5.
Fig. 7. Transcriptional activation of β-catenin target genes is sensitive to E1A and can be rescued by CBP. (A) RT–PCR analyses of siamois expression in whole embryos after injection of RNAs for E1AmRb (0.5 ng), LEFΔN–VP16 (0.1 ng) and CBP (2.0 ng) into the dorsal vegetal zone of four-cell stage embryos as indicated. Control reactions were performed with an RNA sample not treated with reverse transcriptase (–RT). (B) RT–PCR analyses of ectopic siamois induction in animal cap explants by injection of 0.5 ng of β-catS33Amyc RNA with or without 2.0 ng of E1AmCBP or E1AmRb RNA. Control reactions were performed with RNA from total embryos or RNA samples that were not treated with reverse transcriptase (–RT). (C and D) Expression of CBP restores β-catenin target gene induction in the presence of E1AmRb. RT–PCR analyses of siamois and Xnr-3 expression induced in animal cap explants by injection of 0.5 ng of β-catS33Amyc RNA (C) or 40 pg of XWnt-8 RNA (D) with or without 0.5 ng of E1AmRb RNA and 2.0 ng of CBP RNA. In (D), the various RNAs were applied by two consecutive injections to ensure an overlapping spatial protein distribution of E1A or CBP and the secreted XWnt-8. (A–D) For each RNA combination two embryos or two pools of animal cap explants were analyzed separately. EF-1 expression was monitored as control for RNA amount and integrity.
Similar articles
- The C-terminal transactivation domain of beta-catenin is necessary and sufficient for signaling by the LEF-1/beta-catenin complex in Xenopus laevis.
Vleminckx K, Kemler R, Hecht A. Vleminckx K, et al. Mech Dev. 1999 Mar;81(1-2):65-74. doi: 10.1016/s0925-4773(98)00225-1. Mech Dev. 1999. PMID: 10330485 - The ankyrin repeat protein Diversin recruits Casein kinase Iepsilon to the beta-catenin degradation complex and acts in both canonical Wnt and Wnt/JNK signaling.
Schwarz-Romond T, Asbrand C, Bakkers J, Kühl M, Schaeffer HJ, Huelsken J, Behrens J, Hammerschmidt M, Birchmeier W. Schwarz-Romond T, et al. Genes Dev. 2002 Aug 15;16(16):2073-84. doi: 10.1101/gad.230402. Genes Dev. 2002. PMID: 12183362 Free PMC article. - The transcriptional coactivator CBP interacts with beta-catenin to activate gene expression.
Takemaru KI, Moon RT. Takemaru KI, et al. J Cell Biol. 2000 Apr 17;149(2):249-54. doi: 10.1083/jcb.149.2.249. J Cell Biol. 2000. PMID: 10769018 Free PMC article. - Curbing the nuclear activities of beta-catenin. Control over Wnt target gene expression.
Hecht A, Kemler R. Hecht A, et al. EMBO Rep. 2000 Jul;1(1):24-8. doi: 10.1093/embo-reports/kvd012. EMBO Rep. 2000. PMID: 11256619 Free PMC article. Review. - A tight control over Wnt action.
Molenaar M, Destrée O. Molenaar M, et al. Int J Dev Biol. 1999;43(7):675-80. Int J Dev Biol. 1999. PMID: 10668977 Review.
Cited by
- Targeting CBP and p300: Emerging Anticancer Agents.
Masci D, Puxeddu M, Silvestri R, La Regina G. Masci D, et al. Molecules. 2024 Sep 24;29(19):4524. doi: 10.3390/molecules29194524. Molecules. 2024. PMID: 39407454 Free PMC article. Review. - Wnt target gene activation requires β-catenin separation into biomolecular condensates.
Stewart RA, Ding Z, Jeon US, Goodman LB, Tran JJ, Zientko JP, Sabu M, Cadigan KM. Stewart RA, et al. PLoS Biol. 2024 Sep 24;22(9):e3002368. doi: 10.1371/journal.pbio.3002368. eCollection 2024 Sep. PLoS Biol. 2024. PMID: 39316611 Free PMC article. - The role and mechanism of protein post‑translational modification in vascular calcification (Review).
Wang D, Li Q, Xie C. Wang D, et al. Exp Ther Med. 2024 Sep 6;28(5):419. doi: 10.3892/etm.2024.12708. eCollection 2024 Nov. Exp Ther Med. 2024. PMID: 39301258 Free PMC article. Review. - Why Is Wnt/β-Catenin Not Yet Targeted in Routine Cancer Care?
de Pellegars-Malhortie A, Picque Lasorsa L, Mazard T, Granier F, Prévostel C. de Pellegars-Malhortie A, et al. Pharmaceuticals (Basel). 2024 Jul 16;17(7):949. doi: 10.3390/ph17070949. Pharmaceuticals (Basel). 2024. PMID: 39065798 Free PMC article. Review. - The Loss of an Orphan Nuclear Receptor NR2E3 Augments Wnt/β-catenin Signaling via Epigenetic Dysregulation that Enhances Sp1-β catenin-p300 Interactions in Hepatocellular Carcinoma.
Leung YK, Lee SG, Wang J, Guruvaiah P, Rusch NJ, Ho SM, Park C, Kim K. Leung YK, et al. Adv Sci (Weinh). 2024 Aug;11(29):e2308539. doi: 10.1002/advs.202308539. Epub 2024 May 24. Adv Sci (Weinh). 2024. PMID: 38790135 Free PMC article.
References
- Aberle H., Butz,S., Stappert,J., Weissig,H., Kemler,R. and Hoschuetzky,H. (1994) Assembly of the cadherin–catenin complex in vitro with recombinant proteins. J. Cell Sci., 107, 3655–3663. - PubMed
- Aberle H., Schwartz,H. and Kemler,R. (1996) Cadherin–catenin complex: protein interactions and their implications for cadherin function. J. Cell. Biochem., 61, 514–523. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous