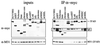Structure-function analysis of SUV39H1 reveals a dominant role in heterochromatin organization, chromosome segregation, and mitotic progression - PubMed (original) (raw)
Structure-function analysis of SUV39H1 reveals a dominant role in heterochromatin organization, chromosome segregation, and mitotic progression
M Melcher et al. Mol Cell Biol. 2000 May.
Abstract
SUV39H1, a human homologue of the Drosophila position effect variegation modifier Su(var)3-9 and of the Schizosaccharomyces pombe silencing factor clr4, encodes a novel heterochromatic protein that transiently accumulates at centromeric positions during mitosis. Using a detailed structure-function analysis of SUV39H1 mutant proteins in transfected cells, we now show that deregulated SUV39H1 interferes at multiple levels with mammalian higher-order chromatin organization. First, forced expression of full-length SUV39H1 (412 amino acids) redistributes endogenous M31 (HP1beta) and induces abundant associations with inter- and metaphase chromatin. These properties depend on the C-terminal SET domain, although the major portion of the SUV39H1 protein (amino acids 89 to 412) does not display affinity for nuclear chromatin. By contrast, the M31 interaction surface, which was mapped to the first 44 N-terminal amino acids, together with the immediately adjacent chromo domain, directs specific accumulation at heterochromatin. Second, cells overexpressing full-length SUV39H1 display severe defects in mitotic progression and chromosome segregation. Surprisingly, whereas localization of centromere proteins is unaltered, the focal, G(2)-specific distribution of phosphorylated histone H3 at serine 10 (phosH3) is dispersed in these cells. This phosH3 shift is not observed with C-terminally truncated mutant SUV39H1 proteins or with deregulated M31. Together, our data reveal a dominant role(s) for the SET domain of SUV39H1 in the distribution of prominent heterochromatic proteins and suggest a possible link between a chromosomal SU(VAR) protein and histone H3.
Figures
FIG. 1
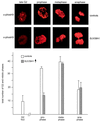
Ectopic SUV39H1 redistributes endogenous M31 (HP1β) in mouse interphase and human metaphase chromatin. (Top panel) Murine Cop8 cells were transiently transfected with a plasmid driving overexpression of (myc)3-tagged full-length SUV39H1 under the control of the CMV promoter-enhancer. Costaining for endogenous M31 and ectopic (myc)3-SUV39H1 was performed with Triton X-100-extracted cells by sequential incubation with (i) rat monoclonal α-M31 antibodies visualized with secondary CY3-conjugated antibodies (red) and (ii) mouse monoclonal α-myc antibodies that were detected with secondary FITC-conjugated antibodies (green). DNA was counterstained with DAPI, which highlights A/T-rich repeat sequences present in the prominent heterochromatic foci (bright blue patches). (Middle and bottom panels) Colocalization of M31 and (myc)3-SUV39H1 on unfixed metaphase-arrested spreads prepared from a “stably” transfected human cell line (HeLa-B3) that overexpresses (myc)3-SUV39H1 in most of the cells (1). Images in the middle panel were taken from a (myc)3-SUV39H1-negative spread, whereas the bottom panel shows the localization of M31 under conditions of (myc)3-SUV39H1 overexpression. Human DNA was counterstained with DA-DAPI. The inserts display the distribution of antigens at enlarged chromosomes.
FIG. 2
Distribution of ectopic M31, full-length SUV39H1, and SUV39H1 mutants in mouse interphase chromatin. Murine Cop8 cells were transiently transfected with overexpression plasmids encoding (myc)3-M31, full-length (myc)3-SUV39H1, and mutant (myc)3-SUV39H1 (indicated to the left; see Fig. 3 and 5 for mutant protein descriptions) and stained by indirect IF with α-myc antibodies (green). DNA was counterstained with DAPI (blue). Transfected cells were either directly processed for IF (left row) or extracted with Triton X-100 to visualize chromatin-associated proteins (right row). After Triton X-100 extraction, only full-length (myc)3-SUV39H1 displays a broad distribution that is not enriched for heterochromatic foci (second panel from the top).
FIG. 3
Distribution of full-length and mutant SUV39H1 on human metaphase chromosome 1. Human HeLa cells were transiently transfected with overexpression plasmids encoding full-length and mutant (myc)3-SUV39H1 (indicated to the left) and stained by indirect IF with α-myc antibodies (green). DNA was counterstained with DA-DAPI (blue). Transfected cells were enriched for metaphase by Colcemid arrest, hypoton treated, spread by cytocentrifugation, and fixed prior to IF. Human chromosomes 1 were visually selected by their large size and prominent block of pericentric heterochromatin. To quantify the intensities of myc signals, chromosomes were also scanned along their entire length (right column).
FIG. 4
In vivo coimmunoprecipitation of mutant SUV39H1 with M31. Human HeLa cells were transiently transfected with overexpression plasmids encoding full-length and mutant (myc)3-SUV39H1 and processed for coimmunoprecipitation (IP) with α-myc antibody beads (right panel). Nuclear extracts were precalibrated and adjusted for comparable amounts of ectopic proteins prior to coimmunoprecipitation (left panel; inputs). Protein blots were probed with α-myc and α-M31 antibodies. The boxed area (marked IgL) indicates residual staining of immunoglobulin light chains that were retained in the precipitated material.
FIG. 5
Summary of mutant SUV39H1 analysis. The names and schematic representations of mutant (myc)3-SUV39H1 are in the leftmost columns. The chromo domain is shown as a grey-shaded box, and the C-terminal SET domain is in black. Numbers refer to amino acid positions in the SUV39H1 protein (1). Mutant proteins are grouped as C-terminal (C-term.) and N-terminal (N-term.) truncations and chromo and SET domain mutations. The names of the two SET domain mutant proteins are abbreviated; they contain short deletions of seven (ΔNHSCDPN323-329) or eight (ΔGEELTFDY358-365) amino acids within the two most-conserved regions of the SET domain (25). The abbreviation nls denotes an SV40 NLS added to the N44 and cysSET mutant proteins. Average expression levels of ectopic proteins were estimated from Western blots of nuclear extracts after transient transfections of Cop8 and HeLa cells, setting the relative abundance of full-length (myc)3-SUV39H1 arbitrarily to 100%. The M31 coimmunoprecipitation (co-IP) data are from Fig. 4, and the M31 coprecipitation potentials are categorized as strong (+++), intermediate (++), weak (+), and negative (−). The data on association with nuclear chromatin are from Fig. 2 and 3. The different distributions in mouse interphase (after Triton X-100 extraction) are indicated as enriched at heterochromatic foci (HET) or broad staining throughout the entire nucleus (dispersed). Association with metaphase chromosomes is summarized as enriched at pericentric heterochromatin (CEN) or broad decoration along the arms (arms). Lack of chromatin association is indicated by a minus sign.
FIG. 6
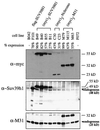
Characterization of stable cell lines. Human HeLa cells were stably cotransfected with a vector conferring G418 resistance and overexpression plasmids encoding full-length flag-SUV39H1, full-length (myc)3-SUV39H1, or (myc)3-Nchromo and (myc)3-M31. Individual cell lines are grouped, with B042 and F072 representing control clones lacking transfected protein. The percentage of cells that stained positive for ectopic protein within a clonal population was determined weekly by indirect IF. The percentage below each cell line designation reflects the respective expression profile of early-passage cells used in the course of this study. Approximately 25 μg of nuclear extracts was processed for immunoblotting with α-myc, α-Suv39h1, and α-M31 antibodies. Positions of endogenous SUV39H1 (48 kDa) and M31 (25 kDa) proteins are indicated.
FIG. 7
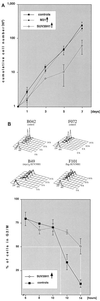
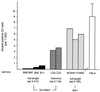
Overexpression of SUV39H1 induces growth retardation and delayed mitotic progression. (A) Cumulative growth curves of three HeLa cell controls (untransfected HeLa, B042, and F072 cells) and three (myc)3-M31 (M79, M115, and M63), one flag-SUV39H1 (F101), and two (myc)3-SUV39H1 (B49 and B40) clones. For each cell line, the total cell numbers were determined in triplicate every other day after reseeding of 105 cells. The graph contains averaged growth curves, and deviations among the clones are indicated by bars. Average generation times for SUV39H1 clones (28.0 h) are approximately 4 or 5 h longer than those for (myc)3-M31-expressing cells (23.7 h) or for the HeLa controls (22.4 h). (B) The three SUV39H1-expressing and three HeLa control cell lines were synchronized at the G1/S boundary by a double thymidine block. Following release, cell cycle progression was monitored by fluorescence-activated cell sorter analysis (top panels; shown for only two cell lines each). The relative percentage of cells in G2/M (i.e., containing 4N DNA) was determined, and progression through G2/M is plotted (averaged graph at the bottom). SUV39H1 clones needed approximately 3 h longer than the HeLa controls to reach a cell population with 50% G2/M cells (indicated by white lines).
FIG. 8
Aberrant chromosome segregation in SUV39H1-overexpressing cells. Logarithmically growing HeLa control cells (B042 and F072) and SUV39H1-expressing clones with a high percentage of overexpressing cells (B40, B49, and F101 cells; Fig. 6) were stained with α-tubulin antibodies (not shown) and DAPI and processed for IF (top panel). SUV39H1 clones display an increased number of cells with aberrant nuclear morphologies (micronuclei and bi- or polunuclei). Many polynuclei also contained multipolar spindles (not shown). At anaphase and telophase, the numbers of chromosomal bridges (including lagging DAPI-stained material between separating sets of chromosomes) are significantly enhanced compared to HeLa control clones. Plotted are the percentages of aberrant structures determined in two independent blind evaluations of ≈500 cells per clone (bottom panel).
FIG. 9
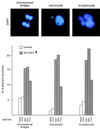
Deregulated SUV39H1 disperses phosH3-positive G2 foci. Logarithmically growing HeLa controls (B042 and F072 cells) and (myc)3-SUV3H1-expressing clones with a high percentage of overexpressing cells (B40 and B49 cells [Fig. 6]) were stained with α-phosH3 antibodies and CY3-conjugated secondary antibodies (red). DNA was counterstained with DAPI (not shown). The pericentric phosH3-positive foci in late G2 (20) and the prominent phosH3 decoration of mitotic chromosomes are shown in the top panel. Evaluation of ≥1,000 cells per (myc)3-SUV39H1-expressing clone only revealed dispersed phosH3 staining in interphase and failed to detect the characteristic phosH3-positive G2 foci. Also plotted are averaged phase indices of ≈200 mitotic cells per clone as determined by DAPI staining and chromosomal morphology (bottom panel).
FIG. 10
Dispersion of phosH3-positive G2 foci is specific for full-length SUV39H1. Logarithmically growing cells of the indicated clones were costained with rabbit polyclonal α-phosH3 and mouse monoclonal α-myc antibodies. Using two independent blind evaluations of ≥1,000 cells per clone, the total numbers of double-positive cells displaying the characteristic phosH3 foci in interphase (Fig. 9) were determined and are plotted. The HeLa bar represents the average number of phosH3-positive G2 foci detected in untransfected HeLa cells and in the two control clones (B042 and F072).
FIG. 11
Modular nature and functional domains of SUV39H1 protein. The 412-aa human SUV39H1 protein contains several conserved regions, including a chromo domain (stippled box), the C-terminal SET domain (black box), SET domain-associated cysteine-rich regions (grey shading), and an N terminus (hatched box) that is shared with Drosophila SU(VAR)3-9 (1). Based on our structure-function analysis, we can assign a direct function to the N-terminal 44 aa and the immediately adjacent chromo domain. The N terminus is an M31 interaction surface which, together with the chromo domain, defines the heterochromatin-targeting region (aa 3 to 118) of SUV39H1. By contrast, the SET domain appears to be functional only in the context of the full-length protein and represents a dominant module that regulates the chromatin association and M31 interaction potentials of SUV39H1. Because mutant SUV39H1 proteins lacking the SET domain do not significantly interfere with the distribution of phosH3-positive G2 foci (Fig. 10), the unexpected link between SUV39H1 and histone H3 is also proposed to be provided by functions of the SET domain. SUV39H1 has recently been shown to be a phosphoprotein with preferred serine phosphorylation of the C-terminal tail, the SET domain, and also, to a lesser extent, the chromo domain (2).
Similar articles
- Functional mammalian homologues of the Drosophila PEV-modifier Su(var)3-9 encode centromere-associated proteins which complex with the heterochromatin component M31.
Aagaard L, Laible G, Selenko P, Schmid M, Dorn R, Schotta G, Kuhfittig S, Wolf A, Lebersorger A, Singh PB, Reuter G, Jenuwein T. Aagaard L, et al. EMBO J. 1999 Apr 1;18(7):1923-38. doi: 10.1093/emboj/18.7.1923. EMBO J. 1999. PMID: 10202156 Free PMC article. - Mitotic phosphorylation of SUV39H1, a novel component of active centromeres, coincides with transient accumulation at mammalian centromeres.
Aagaard L, Schmid M, Warburton P, Jenuwein T. Aagaard L, et al. J Cell Sci. 2000 Mar;113 ( Pt 5):817-29. doi: 10.1242/jcs.113.5.817. J Cell Sci. 2000. PMID: 10671371 - Central role of Drosophila SU(VAR)3-9 in histone H3-K9 methylation and heterochromatic gene silencing.
Schotta G, Ebert A, Krauss V, Fischer A, Hoffmann J, Rea S, Jenuwein T, Dorn R, Reuter G. Schotta G, et al. EMBO J. 2002 Mar 1;21(5):1121-31. doi: 10.1093/emboj/21.5.1121. EMBO J. 2002. PMID: 11867540 Free PMC article. - SU(VAR)3-9 is a conserved key function in heterochromatic gene silencing.
Schotta G, Ebert A, Reuter G. Schotta G, et al. Genetica. 2003 Mar;117(2-3):149-58. doi: 10.1023/a:1022923508198. Genetica. 2003. PMID: 12723694 Review. - Structure, Activity and Function of the Suv39h1 and Suv39h2 Protein Lysine Methyltransferases.
Weirich S, Khella MS, Jeltsch A. Weirich S, et al. Life (Basel). 2021 Jul 16;11(7):703. doi: 10.3390/life11070703. Life (Basel). 2021. PMID: 34357075 Free PMC article. Review.
Cited by
- Centromeric heterochromatin assembly in fission yeast--balancing transcription, RNA interference and chromatin modification.
Alper BJ, Lowe BR, Partridge JF. Alper BJ, et al. Chromosome Res. 2012 Jul;20(5):521-34. doi: 10.1007/s10577-012-9288-x. Chromosome Res. 2012. PMID: 22733402 Free PMC article. Review. - Recruitment of the histone methyltransferase SUV39H1 and its role in the oncogenic properties of the leukemia-associated PML-retinoic acid receptor fusion protein.
Carbone R, Botrugno OA, Ronzoni S, Insinga A, Di Croce L, Pelicci PG, Minucci S. Carbone R, et al. Mol Cell Biol. 2006 Feb;26(4):1288-96. doi: 10.1128/MCB.26.4.1288-1296.2006. Mol Cell Biol. 2006. PMID: 16449642 Free PMC article. - DNA hypermethylation in Drosophila melanogaster causes irregular chromosome condensation and dysregulation of epigenetic histone modifications.
Weissmann F, Muyrers-Chen I, Musch T, Stach D, Wiessler M, Paro R, Lyko F. Weissmann F, et al. Mol Cell Biol. 2003 Apr;23(7):2577-86. doi: 10.1128/MCB.23.7.2577-2586.2003. Mol Cell Biol. 2003. PMID: 12640138 Free PMC article. - The Heterochromatin Protein 1 family.
Lomberk G, Wallrath L, Urrutia R. Lomberk G, et al. Genome Biol. 2006;7(7):228. doi: 10.1186/gb-2006-7-7-228. Genome Biol. 2006. PMID: 17224041 Free PMC article. Review. - Deacetylation and methylation at histone H3 lysine 9 (H3K9) coordinate chromosome condensation during cell cycle progression.
Park JA, Kim AJ, Kang Y, Jung YJ, Kim HK, Kim KC. Park JA, et al. Mol Cells. 2011 Apr;31(4):343-9. doi: 10.1007/s10059-011-0044-4. Epub 2011 Feb 2. Mol Cells. 2011. PMID: 21359677 Free PMC article.
References
- Aagaard L, Laible G, Selenko P, Schmid M, Dorn R, Schotta G, Kuhfittig S, Wolf A, Lebersorger A, Singh P B, Reuter G, Jenuwein T. Functional mammalian homologues of the Drosophila PEV modifier Su(var)3-9 encode centromere-associated proteins which complex with the heterochromatin component M31. EMBO J. 1999;18:1923–1938. - PMC - PubMed
- Aagaard L, Schmid M, Warburton P, Jenuwein T. Mitotic phosphorylation of SUV39H1, a novel component of active centromeres, coincides with transient accumulation at mammalian centromeres. J Cell Sci. 2000;113:817–829. - PubMed
- Allshire R C, Nimmo E R, Ekwall K, Javerzat J P, Granston G. Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev. 1995;9:218–233. - PubMed
- Baksa K, Morawietz H, Dombradi V, Axton M, Taubert H, Szabo G, Török I, Udvardy A, Gyuorkovics H, Szöör B, Glover D, Reuter G, Gausz J. Mutations in the protein phosphatase 1 gene at 87B can differentially affect suppression of position effect variegation and mitosis in Drosophila melanogaster. Genetics. 1993;135:117–125. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources