A microsphere-based assay for multiplexed single nucleotide polymorphism analysis using single base chain extension - PubMed (original) (raw)
A microsphere-based assay for multiplexed single nucleotide polymorphism analysis using single base chain extension
J Chen et al. Genome Res. 2000 Apr.
Abstract
A rapid, high throughput readout for single-nucleotide polymorphism (SNP) analysis was developed employing single base chain extension and cytometric analysis of an array of fluorescent microspheres. An array of fluorescent microspheres was coupled with uniquely identifying sequences, termed complementary ZipCodes (cZipCodes), which allowed for multiplexing possibilities. For a given assay, querying a polymorphic base involved extending an oligonucleotide containing both a ZipCode and a SNP-specific sequence with a DNA polymerase and a pair of fluoresceinated dideoxynucleotides. To capture the reaction products for analysis, the ZipCode portion of the oligonucleotide was hybridized with its cZipCodes on the microsphere. Flow cytometry was used for microsphere decoding and SNP typing by detecting the fluorescein label captured on the microspheres. In addition to multiplexing capability, the ZipCode system allows multiple sets of SNPs to be analyzed by a limited set of cZipCode-attached microspheres. A standard set of non-cross reactive ZipCodes was established experimentally and the accuracy of the system was validated by comparison with genotypes determined by other technologies. From a total of 58 SNPs, 55 SNPs were successfully analyzed in the first pass using this assay format and all 181 genotypes across the 55 SNPs were correct. These data demonstrate that the microsphere-based single base chain extension (SBCE) method is a sensitive and reliable assay. It can be readily adapted to an automated, high-throughput genotyping system. [Primer sequences used in this study are available as online supplementary materials at www.genome.org.\]
Figures
Figure 1
Schematic presentation of the microsphere-based single base chain extension assays. DNA fragments containing the polymorphic site to be typed were amplified either individually or by multiplexed PCR (step 1). The PCR products containing a SNP site were pooled and treated with SAP and exonuclease I (step 2). After heat inactivation of the enzymes, the PCR products were used in the SBCE reactions (step 3) as described in Methods. For every SNP, one capture oligonucleotide probe with a unique ZipCode sequence was designed and used to assay the two alleles in each of two separate wells with a different labeled ddNTP per well. Multiplexed SNP analysis could be achieved by the employment of different ZipCode sequences for different SNPs in the presence of pooled PCR products. After the completion of the SBCE reaction, ∼1200 of each type of microsphere [with an attached oligonucleotide encoding the complement to the ZipCode sequences and a common luciferase sequence (SeqLUC)] were added to the completed SBCE reactions. The hybridization reactions were carried out at 40°C in the presence of NaCl for >2 hr (step 4). The microspheres were then subjected to flow cytometric analysis (step 5). Minimums of 100 of each type of microsphere were read and the mean value of MESF was used for determining the genotypes. The fluorescence signal of the corresponding microsphere without SBCE reactions (microsphere alone) or SBCE reactions without AmpliTaq FS were subtracted from the MESF values.
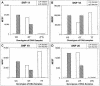
Figure 2
SNP analysis in multiplexed reactions. PCR products were amplified individually from genomic DNA with either homozygous genotypes (CC and TT) or heterozygous genotype (CT). PCR products were then pooled according to their known genotypes into three separate groups. For example, one pool contained the homozygous PCR products (CC) from the four SNPs and so on. The three pools (15 ng of each PCR products) were used as templates and assayed separately for either A or G (striped columns) incorporation with the antisense probe as described in Methods. The MESF values from each of the three genotypes for each SNP are grouped together and shown. About 10,000 microspheres were pretreated with BSA at 1 mg/ml for 45 min and then added to each reaction to capture the SBCE products. The fluorescent intensity of the microspheres is represented by the MESF values on the _y_-axis. A pair of numbers from the A and G reactions determine the genotypes of the samples analyzed. Genotypes of the DNA samples were labeled as CC, CT, and TT. The absence of the PCR products for SNP11 and SNP20 for the TT allele is indicated by (TT).
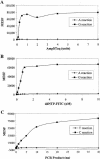
Figure 3
Optimization of SBCE reactions. The MESF values in the _y_-axis represent the mean fluorescence per microsphere. (A) Titration of the AmpliTaq FS enzyme for SNP18 in a multiplex reaction of four SNPs as used in Figure 2. A mixture of 15 ng PCR products containing homozygous CC genotypes was used as template for either G (█) or A (♦) reactions and the assays were performed with the anti-sense probe as described in Methods. A total of 10,000 microspheres were used for each reaction. The value obtained from the reaction in the absence of enzyme was subtracted from the data points. (B) Titration of ddNTPs for SNP18 was done using the same multiplex conditions as in (A). The experiments were performed with various amounts of fluorescent-labeled ddNTP. The ratio of labeled to unlabeled ddNTPs was kept constant at 1:3. The signals remained fairly consistent for the G reactions (█,) at and above 0.75 n
m
of ddGTP; A reactions (♦) remained near 0. The results for the other three SNPs are nearly identical to the results shown here. (C) Effect of PCR products on the enzymatic activities. A PCR product (250 bp) generated from a homozygous (CC) DNA sample for SNP18 was used as template for assaying the incorporation of either C (█) or T (♦) nucleotides with the sense capture oligonucleotide.
Similar articles
- Fluorescent microsphere-based readout technology for multiplexed human single nucleotide polymorphism analysis and bacterial identification.
Ye F, Li MS, Taylor JD, Nguyen Q, Colton HM, Casey WM, Wagner M, Weiner MP, Chen J. Ye F, et al. Hum Mutat. 2001 Apr;17(4):305-16. doi: 10.1002/humu.28. Hum Mutat. 2001. PMID: 11295829 - Multiplexed single nucleotide polymorphism genotyping by oligonucleotide ligation and flow cytometry.
Iannone MA, Taylor JD, Chen J, Li MS, Rivers P, Slentz-Kesler KA, Weiner MP. Iannone MA, et al. Cytometry. 2000 Feb 1;39(2):131-40. Cytometry. 2000. PMID: 10679731 - Flow cytometric platform for high-throughput single nucleotide polymorphism analysis.
Taylor JD, Briley D, Nguyen Q, Long K, Iannone MA, Li MS, Ye F, Afshari A, Lai E, Wagner M, Chen J, Weiner MP. Taylor JD, et al. Biotechniques. 2001 Mar;30(3):661-6, 668-9. doi: 10.2144/01303dd04. Biotechniques. 2001. PMID: 11252801 - Multiplexed microsphere-based flow cytometric assays.
Kellar KL, Iannone MA. Kellar KL, et al. Exp Hematol. 2002 Nov;30(11):1227-37. doi: 10.1016/s0301-472x(02)00922-0. Exp Hematol. 2002. PMID: 12423675 Review. - Medium- to high-throughput SNP genotyping using VeraCode microbeads.
Lin CH, Yeakley JM, McDaniel TK, Shen R. Lin CH, et al. Methods Mol Biol. 2009;496:129-42. doi: 10.1007/978-1-59745-553-4_10. Methods Mol Biol. 2009. PMID: 18839109 Review.
Cited by
- Novel single nucleotide polymorphisms in the heat shock protein 70.1 gene in South African Nguni crossbred cattle.
Mkize LS, Zishiri OT. Mkize LS, et al. Trop Anim Health Prod. 2020 Mar;52(2):893-901. doi: 10.1007/s11250-019-02088-6. Epub 2019 Oct 23. Trop Anim Health Prod. 2020. PMID: 31643021 - Multiplex Detection of Five Canine Viral Pathogens for Dogs as Laboratory Animals by the Luminex xTAG Assay.
Wu M, Cong F, Zhu Y, Lian Y, Chen M, Huang R, Guo P. Wu M, et al. Front Microbiol. 2018 Aug 17;9:1783. doi: 10.3389/fmicb.2018.01783. eCollection 2018. Front Microbiol. 2018. PMID: 30174654 Free PMC article. - Multiphoton-Polymerized 3D Protein Assay.
Wollhofen R, Axmann M, Freudenthaler P, Gabriel C, Röhrl C, Stangl H, Klar TA, Jacak J. Wollhofen R, et al. ACS Appl Mater Interfaces. 2018 Jan 17;10(2):1474-1479. doi: 10.1021/acsami.7b13183. Epub 2018 Jan 5. ACS Appl Mater Interfaces. 2018. PMID: 29280613 Free PMC article. - xMAP Technology: Applications in Detection of Pathogens.
Reslova N, Michna V, Kasny M, Mikel P, Kralik P. Reslova N, et al. Front Microbiol. 2017 Jan 25;8:55. doi: 10.3389/fmicb.2017.00055. eCollection 2017. Front Microbiol. 2017. PMID: 28179899 Free PMC article. Review. - Detection of first-line anti-tuberculosis drug resistance mutations by allele-specific primer extension on a microsphere-based platform.
Lee SH, Choi HB, Yu SY, Chang UJ, Kim CK, Kim HJ. Lee SH, et al. Ann Lab Med. 2015 Sep;35(5):487-93. doi: 10.3343/alm.2015.35.5.487. Ann Lab Med. 2015. PMID: 26206684 Free PMC article.
References
- Cooper DN, Smith BA, Cooke HJ, Niemann S, Schmidtke J. An estimate of unique DNA sequence heterozygosity in the human genome. Hum Genet. 1985;69:201–205. - PubMed
- Fu DJ, Tang K, Braun A, Reuter D, Darnhofer-Demar B, Little DP, O'Donnell MJ, Cantor CR, Koster H. Sequencing exons 5 to 8 of the p53 gene by MALDI-TOF mass spectrometry. Nat Biotechnol. 1998;16:381–384. - PubMed
- Fulton RJ, McDade RL, Smith PL, Kienker LJ, Kettman, J. JR. Advanced multiplexed analysis with the FlowMetrix system. Clin Chem. 1997;43:1749–1756. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials