Evolution of Antp-class genes and differential expression of Hydra Hox/paraHox genes in anterior patterning - PubMed (original) (raw)
Evolution of Antp-class genes and differential expression of Hydra Hox/paraHox genes in anterior patterning
D Gauchat et al. Proc Natl Acad Sci U S A. 2000.
Abstract
The conservation of developmental functions exerted by Antp-class homeoproteins in protostomes and deuterostomes suggested that homologs with related functions are present in diploblastic animals. Our phylogenetic analyses showed that Antp-class homeodomains belong either to non-Hox or to Hox/paraHox families. Among the 13 non-Hox families, 9 have diploblastic homologs, Msx, Emx, Barx, Evx, Tlx, NK-2, and Prh/Hex, Not, and Dlx, reported here. Among the Hox/paraHox, poriferan sequences were not found, and the cnidarian sequences formed at least five distinct cnox families. Two are significantly related to the paraHox Gsx (cnox-2) and the mox (cnox-5) sequences, whereas three display some relatedness to the Hox paralog groups 1 (cnox-1), 9/10 (cnox-3) and the paraHox cdx (cnox-4). Intermediate Hox/paraHox genes (PG 3 to 8 and lox) did not have clear cnidarian counterparts. In Hydra, cnox-1, cnox-2, and cnox-3 were not found chromosomally linked within a 150-kb range and displayed specific expression patterns in the adult head. During regeneration, cnox-1 was expressed as an early gene whatever the polarity, whereas cnox-2 was up-regulated later during head but not foot regeneration. Finally, cnox-3 expression was reestablished in the adult head once it was fully formed. These results suggest that the Hydra genes related to anterior Hox/paraHox genes are involved at different stages of apical differentiation. However, the positional information defining the oral/aboral axis in Hydra cannot be correlated strictly to that characterizing the anterior-posterior axis in vertebrates or arthropods.
Figures
Figure 1
Conserved residues at specific positions provide a common signature for all Antp-class HDs and thonon-Hox or the Hox/paraHox HDs (bold). In addition, a family-specific signature is given where identical or equivalent residues between diploblastic and bilaterian sequences are underlined.
Figure 2
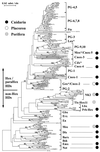
Phylogenetic relationships between 200 Antp-class genes inferred by NJ analysis by using Dayhoff's PAM distance matrix. The tree was rooted with a Prd-class sequence. All branch lengths were drawn to scale. Numbers at nodes indicate percentages of 200 bootstrap replicates that support the branch; values under 50% are omitted, except for some significantly important nodes. Support values for each family defined on the right are marked in bold. Sequences from diploblasts are in shaded boxes.
Figure 3
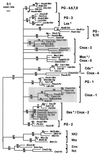
Phylogenetic tree of 57 Hox/paraHox HD sequences inferred by ML analysis where 242,075 of the 677,040 possible quartets of sequences (35.8%) were unresolved, leading to a multifurcating tree with a Log likelihood value of −3,656.89. Numbers above the branches correspond to the quartet puzzling support values; numbers under branches indicate percentages of 1,000 bootstrap replicates for NJ analysis of the same dataset. Ten sequences from non-Hox families (NK2, Msx, Not, and Emx) were used as an outgroup. Shaded boxes as in Fig. 2.
Figure 4
PFGE analysis of Cv genomic DNA predigested with _Cla_I (2), NruI (3), MluI (4), NarI (5), _Pvu_I (6), _Sma_I (7), or undigested (1) and hybridized to cnox-1 Cv, cnox-2 Cv, cnox-3 Hv, and Bar-H1 Cv [initially named cnox-3 Cv (15)] probes.
Figure 5
Expression pattern of the Hv Hox/paraHox genes, cnox-1 (Top), cnox-2 (Middle), and cnox-3 (Bottom) in adult (Left) and regenerating Hydra. Time points after cutting are given; arrowheads and arrows indicate foot- and head-regenerating stumps, respectively. st, budding stage.
Figure 6
Scheme describing a plausible scenario of the evolution of Antp-class genes from diploblastic to bilaterian animals.
Similar articles
- Evolution of Hox-like genes in Cnidaria: Study of Hydra Hox repertoire reveals tailor-made Hox-code for Cnidarians.
Reddy PC, Unni MK, Gungi A, Agarwal P, Galande S. Reddy PC, et al. Mech Dev. 2015 Nov;138 Pt 2:87-96. doi: 10.1016/j.mod.2015.08.005. Epub 2015 Aug 13. Mech Dev. 2015. PMID: 26278345 - Homeobox genes in the cnidarian Eleutheria dichotoma: evolutionary implications for the origin of Antennapedia-class (HOM/Hox) genes.
Kuhn K, Streit B, Schierwater B. Kuhn K, et al. Mol Phylogenet Evol. 1996 Aug;6(1):30-8. doi: 10.1006/mpev.1996.0055. Mol Phylogenet Evol. 1996. PMID: 8812303 - Expression of a Gsx parahox gene, Cnox-2, in colony ontogeny in Hydractinia (Cnidaria: Hydrozoa).
Cartwright P, Schierwater B, Buss LW. Cartwright P, et al. J Exp Zool B Mol Dev Evol. 2006 Sep 15;306(5):460-9. doi: 10.1002/jez.b.21106. J Exp Zool B Mol Dev Evol. 2006. PMID: 16615106 - Evolution of homeobox genes.
Holland PW. Holland PW. Wiley Interdiscip Rev Dev Biol. 2013 Jan-Feb;2(1):31-45. doi: 10.1002/wdev.78. Epub 2012 Sep 10. Wiley Interdiscip Rev Dev Biol. 2013. PMID: 23799629 Review. - Hox and ParaHox genes: a review on molluscs.
Biscotti MA, Canapa A, Forconi M, Barucca M. Biscotti MA, et al. Genesis. 2014 Dec;52(12):935-45. doi: 10.1002/dvg.22839. Epub 2014 Nov 28. Genesis. 2014. PMID: 25394269 Review.
Cited by
- The canonical Wnt signaling activator, R-spondin2, regulates craniofacial patterning and morphogenesis within the branchial arch through ectodermal-mesenchymal interaction.
Jin YR, Turcotte TJ, Crocker AL, Han XH, Yoon JK. Jin YR, et al. Dev Biol. 2011 Apr 1;352(1):1-13. doi: 10.1016/j.ydbio.2011.01.004. Epub 2011 Jan 13. Dev Biol. 2011. PMID: 21237142 Free PMC article. - Msx1 and Msx2 function together in the regulation of primordial germ cell migration in the mouse.
Sun J, Ting MC, Ishii M, Maxson R. Sun J, et al. Dev Biol. 2016 Sep 1;417(1):11-24. doi: 10.1016/j.ydbio.2016.07.013. Epub 2016 Jul 18. Dev Biol. 2016. PMID: 27435625 Free PMC article. - Beyond the Hox: how widespread is homeobox gene clustering?
Holland PW. Holland PW. J Anat. 2001 Jul-Aug;199(Pt 1-2):13-23. doi: 10.1046/j.1469-7580.2001.19910013.x. J Anat. 2001. PMID: 11523814 Free PMC article. Review. - The cnidarian-bilaterian ancestor possessed at least 56 homeoboxes: evidence from the starlet sea anemone, Nematostella vectensis.
Ryan JF, Burton PM, Mazza ME, Kwong GK, Mullikin JC, Finnerty JR. Ryan JF, et al. Genome Biol. 2006;7(7):R64. doi: 10.1186/gb-2006-7-7-R64. Genome Biol. 2006. PMID: 16867185 Free PMC article. - Gene Loss may have Shaped the Cnidarian and Bilaterian Hox and ParaHox Complement.
Steinworth BM, Martindale MQ, Ryan JF. Steinworth BM, et al. Genome Biol Evol. 2023 Jan 4;15(1):evac172. doi: 10.1093/gbe/evac172. Genome Biol Evol. 2023. PMID: 36508343 Free PMC article.
References
- Lewis E B. Nature (London) 1978;276:565–570. - PubMed
- Gehring W J. Cell. 1985;40:3–5. - PubMed
- Graham A, Papalopulu N, Krumlauf R. Cell. 1989;57:367–378. - PubMed
- Simeone A, Acampora D, Gulisano M, Stornaiuolo A, Boncinelli E. Nature (London) 1992;358:687–690. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources