Robust perfect adaptation in bacterial chemotaxis through integral feedback control - PubMed (original) (raw)
Robust perfect adaptation in bacterial chemotaxis through integral feedback control
T M Yi et al. Proc Natl Acad Sci U S A. 2000.
Abstract
Integral feedback control is a basic engineering strategy for ensuring that the output of a system robustly tracks its desired value independent of noise or variations in system parameters. In biological systems, it is common for the response to an extracellular stimulus to return to its prestimulus value even in the continued presence of the signal-a process termed adaptation or desensitization. Barkai, Alon, Surette, and Leibler have provided both theoretical and experimental evidence that the precision of adaptation in bacterial chemotaxis is robust to dramatic changes in the levels and kinetic rate constants of the constituent proteins in this signaling network [Alon, U., Surette, M. G., Barkai, N. & Leibler, S. (1998) Nature (London) 397, 168-171]. Here we propose that the robustness of perfect adaptation is the result of this system possessing the property of integral feedback control. Using techniques from control and dynamical systems theory, we demonstrate that integral control is structurally inherent in the Barkai-Leibler model and identify and characterize the key assumptions of the model. Most importantly, we argue that integral control in some form is necessary for a robust implementation of perfect adaptation. More generally, integral control may underlie the robustness of many homeostatic mechanisms.
Figures
Figure 1
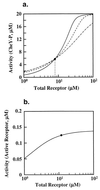
Robust and nonrobust perfect adaptation. The dependence of steady-state system activity on total receptor concentration was calculated by using equilibrium analysis for three concentrations of chemoattractant: (i) L = 0 (solid), (ii) L = 1 μM (dashed), and (iii) L = 1 mM (dashed–dot). The filled circle indicates the value of total receptor used in the model. (a) Spiro model. The system activity is measured in terms of the concentration of CheY-P. The intersection of the three lines, which coincides with the filled circle, represents the value of total receptor at which perfect adaptation exists. (b) Barkai–Leibler model. The system activity is measured in terms of the concentration of active receptor complexes. The three lines completely overlap. These plots were performed by using the program
xppaut
(34, 35).
Figure 2
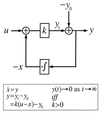
A block diagram of integral feedback control. The variable_u_ is the input for a process with gain k. The difference between the actual output y_1 and the steady-state output y_0 represents the normalized output or error, y. Integral control arises through the feedback loop in which the time integral of_y, x, is fed back into the system. As a result, we have ẋ = y and_y = 0 at steady-state for all u. In the Barkai–Leibler model of the bacterial chemotaxis signaling system, the chemoattractant is the input, receptor activity is the output, and −x approximates the methylation level of the receptors.
Figure 3
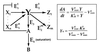
Robust regulation of the steady-state level of a pathway component. In this hypothetical biochemical pathway, the level of the component_Y_ is maintained at a constant steady-state level by an integral feedback loop mediated by the regulator molecule_A_ and the enzymes that produce and remove_A_, _E_3 and_E_4. Both the upstream molecules (Xi) and enzymes (E_1_i) and the downstream molecules (Zi) and enzymes (E_2_i) do not influence the steady-state concentration of Y,_Y_0, assuming that the system is stable. The equation for _Y_0 was derived by using Michaelis–Menten kinetics and the assumption that_E_4 is operating at saturation.V max_3 and_V max_4 are the maximal velocities, and_K m_3 and_K _m_4 are the Michaelis constants for _E_3 and_E_4.
Comment in
- Cell signaling pathways as control modules: complexity for simplicity?
Lauffenburger DA. Lauffenburger DA. Proc Natl Acad Sci U S A. 2000 May 9;97(10):5031-3. doi: 10.1073/pnas.97.10.5031. Proc Natl Acad Sci U S A. 2000. PMID: 10805765 Free PMC article. No abstract available.
Similar articles
- Perfect and near-perfect adaptation in a model of bacterial chemotaxis.
Mello BA, Tu Y. Mello BA, et al. Biophys J. 2003 May;84(5):2943-56. doi: 10.1016/S0006-3495(03)70021-6. Biophys J. 2003. PMID: 12719226 Free PMC article. - Ultrasensitivity and fluctuations in the Barkai-Leibler model of chemotaxis receptors in Escherichia coli.
Roy U, Gopalakrishnan M. Roy U, et al. PLoS One. 2017 Apr 13;12(4):e0175309. doi: 10.1371/journal.pone.0175309. eCollection 2017. PLoS One. 2017. PMID: 28406996 Free PMC article. - A universal biomolecular integral feedback controller for robust perfect adaptation.
Aoki SK, Lillacci G, Gupta A, Baumschlager A, Schweingruber D, Khammash M. Aoki SK, et al. Nature. 2019 Jun;570(7762):533-537. doi: 10.1038/s41586-019-1321-1. Epub 2019 Jun 19. Nature. 2019. PMID: 31217585 - Adaptive molecular networks controlling chemotactic migration: dynamic inputs and selection of the network architecture.
Chang H, Levchenko A. Chang H, et al. Philos Trans R Soc Lond B Biol Sci. 2013 Sep 23;368(1629):20130117. doi: 10.1098/rstb.2013.0117. Print 2013. Philos Trans R Soc Lond B Biol Sci. 2013. PMID: 24062588 Free PMC article. Review. - Overview of mathematical approaches used to model bacterial chemotaxis I: the single cell.
Tindall MJ, Porter SL, Maini PK, Gaglia G, Armitage JP. Tindall MJ, et al. Bull Math Biol. 2008 Aug;70(6):1525-69. doi: 10.1007/s11538-008-9321-6. Epub 2008 Jul 19. Bull Math Biol. 2008. PMID: 18642048 Review.
Cited by
- Signaling, Deconstructed: Using Optogenetics to Dissect and Direct Information Flow in Biological Systems.
Farahani PE, Reed EH, Underhill EJ, Aoki K, Toettcher JE. Farahani PE, et al. Annu Rev Biomed Eng. 2021 Jul 13;23:61-87. doi: 10.1146/annurev-bioeng-083120-111648. Epub 2021 Mar 15. Annu Rev Biomed Eng. 2021. PMID: 33722063 Free PMC article. Review. - A dynamical feedback model for adaptation in the olfactory transduction pathway.
De Palo G, Boccaccio A, Miri A, Menini A, Altafini C. De Palo G, et al. Biophys J. 2012 Jun 20;102(12):2677-86. doi: 10.1016/j.bpj.2012.04.040. Epub 2012 Jun 19. Biophys J. 2012. PMID: 22735517 Free PMC article. - Chemoattractant signaling in dictyostelium: adaptation and amplification.
Iglesias PA. Iglesias PA. Sci Signal. 2012 Feb 28;5(213):pe8. doi: 10.1126/scisignal.2002897. Sci Signal. 2012. PMID: 22375054 Free PMC article. Review. - Characterization of the adaptation module of the signaling network in bacterial chemotaxis by measurement of step responses.
Yuan J, Berg HC. Yuan J, et al. Biophys J. 2012 Sep 19;103(6):L31-3. doi: 10.1016/j.bpj.2012.08.019. Biophys J. 2012. PMID: 22995514 Free PMC article. - The nanotechnology of life-inspired systems.
Grzybowski BA, Huck WT. Grzybowski BA, et al. Nat Nanotechnol. 2016 Jul 6;11(7):585-92. doi: 10.1038/nnano.2016.116. Nat Nanotechnol. 2016. PMID: 27380745
References
- Stryer L. Biochemistry. New York: Freeman; 1995.
- Berg H C, Brown D A. Nature (London) 1972;239:500–504. - PubMed
- Stock J B, Lukat G S, Stock A M. Annu Rev Biophys Biophys Chem. 1991;20:109–136. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources