Metabolic analysis of Escherichia coli in the presence and absence of the carboxylating enzymes phosphoenolpyruvate carboxylase and pyruvate carboxylase - PubMed (original) (raw)
Metabolic analysis of Escherichia coli in the presence and absence of the carboxylating enzymes phosphoenolpyruvate carboxylase and pyruvate carboxylase
R R Gokarn et al. Appl Environ Microbiol. 2000 May.
Abstract
Fermentation patterns of Escherichia coli with and without the phosphoenolpyruvate carboxylase (PPC) and pyruvate carboxylase (PYC) enzymes were compared under anaerobic conditions with glucose as a carbon source. Time profiles of glucose and fermentation product concentrations were determined and used to calculate metabolic fluxes through central carbon pathways during exponential cell growth. The presence of the Rhizobium etli pyc gene in E. coli (JCL1242/pTrc99A-pyc) restored the succinate producing ability of E. coli ppc null mutants (JCL1242), with PYC competing favorably with both pyruvate formate lyase and lactate dehydrogenase. Succinate formation was slightly greater by JCL1242/pTrc99A-pyc than by cells which overproduced PPC (JCL1242/pPC201, ppc(+)), even though PPC activity in cell extracts of JCL1242/pPC201 (ppc(+)) was 40-fold greater than PYC activity in extracts of JCL1242/pTrc99a-pyc. Flux calculations indicate that during anaerobic metabolism the pyc(+) strain had a 34% greater specific glucose consumption rate, a 37% greater specific rate of ATP formation, and a 6% greater specific growth rate compared to the ppc(+) strain. In light of the important position of pyruvate at the juncture of NADH-generating pathways and NADH-dissimilating branches, the results show that when PPC or PYC is expressed, the metabolic network adapts by altering the flux to lactate and the molar ratio of ethanol to acetate formation.
Figures
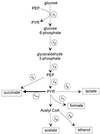
FIG. 1
Fermentative pathways of E. coli grown in a glucose-limited rich medium. This figure depicts the principal branches of glucose metabolism by E. coli under anaerobic conditions. PYR, pyruvate.
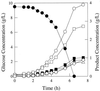
FIG. 2
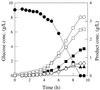
Fermentation pattern of E. coli strain VJS676 growing in glucose-limited rich media. Symbols: ●, glucose; ■, succinate; ▴, lactate; ○, formate; □, acetate; ▵, ethanol; ◊, biomass.
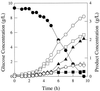
FIG. 3
Fermentation pattern of E. coli strain JCL1242 (ppc null mutant) growing in glucose-limited rich medium. Symbols: ●, glucose; ■, succinate; ▴, lactate; ○, formate; □, acetate; ▵, ethanol; ◊, biomass.
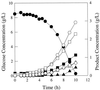
FIG. 4
Fermentation pattern of E. coli strain JCL1242/pPC201 (ppc+) growing in glucose-limited rich medium. Symbols: ●, glucose; ■, succinate; ▴, lactate; ○, formate; □, acetate; ▵, ethanol, and ◊, biomass.
FIG. 5
Fermentation pattern of E. coli strain JCL1242/pTrc99A-pyc growing in glucose-limited rich medium. Symbols: ●, glucose; ■, succinate; ▴, lactate; ○, formate; □, acetate; ▵, ethanol; ◊, biomass.
Similar articles
- The physiological effects and metabolic alterations caused by the expression of Rhizobium etli pyruvate carboxylase in Escherichia coli.
Gokarn RR, Evans JD, Walker JR, Martin SA, Eiteman MA, Altman E. Gokarn RR, et al. Appl Microbiol Biotechnol. 2001 Jul;56(1-2):188-95. doi: 10.1007/s002530100661. Appl Microbiol Biotechnol. 2001. PMID: 11499929 - Effects of growth mode and pyruvate carboxylase on succinic acid production by metabolically engineered strains of Escherichia coli.
Vemuri GN, Eiteman MA, Altman E. Vemuri GN, et al. Appl Environ Microbiol. 2002 Apr;68(4):1715-27. doi: 10.1128/AEM.68.4.1715-1727.2002. Appl Environ Microbiol. 2002. PMID: 11916689 Free PMC article. - Effect of Sorghum vulgare phosphoenolpyruvate carboxylase and Lactococcus lactis pyruvate carboxylase coexpression on succinate production in mutant strains of Escherichia coli.
Lin H, San KY, Bennett GN. Lin H, et al. Appl Microbiol Biotechnol. 2005 Jun;67(4):515-23. doi: 10.1007/s00253-004-1789-x. Epub 2004 Nov 24. Appl Microbiol Biotechnol. 2005. PMID: 15565333 - Anaplerotic function of phosphoenolpyruvate carboxylase in Bradyrhizobium japonicum USDA110.
Dunn MF. Dunn MF. Curr Microbiol. 2011 Jun;62(6):1782-8. doi: 10.1007/s00284-011-9928-y. Epub 2011 Apr 10. Curr Microbiol. 2011. PMID: 21479798 - Alteration of the biochemical valves in the central metabolism of Escherichia coli.
Liao JC, Chao YP, Patnaik R. Liao JC, et al. Ann N Y Acad Sci. 1994 Nov 30;745:21-34. doi: 10.1111/j.1749-6632.1994.tb44361.x. Ann N Y Acad Sci. 1994. PMID: 7832509 Review.
Cited by
- The Interaction between Human Microbes and Advanced Glycation End Products: The Role of Klebsiella X15 on Advanced Glycation End Products' Degradation.
Shi A, Ji X, Li W, Dong L, Wu Y, Zhang Y, Liu X, Zhang Y, Wang S. Shi A, et al. Nutrients. 2024 Mar 6;16(5):754. doi: 10.3390/nu16050754. Nutrients. 2024. PMID: 38474882 Free PMC article. - Conversion of Escherichia coli into Mixotrophic CO2 Assimilation with Malate and Hydrogen Based on Recombinant Expression of 2-Oxoglutarate:Ferredoxin Oxidoreductase Using Adaptive Laboratory Evolution.
Cheng YC, Huang WH, Lo SC, Huang E, Chiang EI, Huang CC, Yang YT. Cheng YC, et al. Microorganisms. 2023 Jan 19;11(2):253. doi: 10.3390/microorganisms11020253. Microorganisms. 2023. PMID: 36838218 Free PMC article. - Hydrolysis of lignocellulose to succinic acid: a review of treatment methods and succinic acid applications.
Zhou S, Zhang M, Zhu L, Zhao X, Chen J, Chen W, Chang C. Zhou S, et al. Biotechnol Biofuels Bioprod. 2023 Jan 2;16(1):1. doi: 10.1186/s13068-022-02244-5. Biotechnol Biofuels Bioprod. 2023. PMID: 36593503 Free PMC article. Review. - Biosynthetic Pathway and Metabolic Engineering of Succinic Acid.
Liu X, Zhao G, Sun S, Fan C, Feng X, Xiong P. Liu X, et al. Front Bioeng Biotechnol. 2022 Mar 8;10:843887. doi: 10.3389/fbioe.2022.843887. eCollection 2022. Front Bioeng Biotechnol. 2022. PMID: 35350186 Free PMC article. Review. - Accelerated Electro-Fermentation of Acetoin in Escherichia coli by Identifying Physiological Limitations of the Electron Transfer Kinetics and the Central Metabolism.
Beblawy S, Philipp LA, Gescher J. Beblawy S, et al. Microorganisms. 2020 Nov 23;8(11):1843. doi: 10.3390/microorganisms8111843. Microorganisms. 2020. PMID: 33238546 Free PMC article.
References
- Aiba S, Matsuoka M. Identification of metabolic model: citrate production from glucose by Candida lipolytica. Biotechnol Bioeng. 1979;21:1373–1386.
- Amann E, Ochs B, Abel K-J. Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene. 1988;69:301–315. - PubMed
- Attwood P V. The structure and the mechanism of action of pyruvate carboxylase. Int J Biochem Cell Biol. 1995;27:231–249. - PubMed
- Blaschkowski H P, Neuer G, Ludwig-Festl M, Knappe J. Routes of flavodoxin and ferredoxin reduction in E. coli. Eur J Biochem. 1980;123:563–569. - PubMed
- Bunch P K, Mat-Jan F, Lee N, Clark D P. The ldhA gene encoding the fermentative lactate dehydrogenase of Escherichia coli. Microbiology. 1997;143:187–195. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases