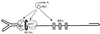Cyclin A activates the DNA polymerase delta -dependent elongation machinery in vitro: A parvovirus DNA replication model - PubMed (original) (raw)
Cyclin A activates the DNA polymerase delta -dependent elongation machinery in vitro: A parvovirus DNA replication model
T Bashir et al. Proc Natl Acad Sci U S A. 2000.
Abstract
Replication of the single-stranded linear DNA genome of parvovirus minute virus of mice (MVM) starts with complementary strand synthesis from the 3'-terminal snap-back telomere, which serves as a primer for the formation of double-stranded replicative form (RF) DNA. This DNA elongation reaction, designated conversion, is exclusively dependent on cellular factors. In cell extracts, we found that complementary strand synthesis was inhibited by the cyclin-dependent kinase inhibitor p21(WAF1/CIP1) and rescued by the addition of proliferating cell nuclear antigen, arguing for the involvement of DNA polymerase (Pol) delta in the conversion reaction. In vivo time course analyses using synchronized MVM-infected A9 cells allowed initial detection of MVM RF DNA at the G(1)/S phase transition, coinciding with the onset of cyclin A expression and cyclin A-associated kinase activity. Under in vitro conditions, formation of RF DNA was efficiently supported by A9 S cell extracts, but only marginally by G(1) cell extracts. Addition of recombinant cyclin A stimulated DNA conversion in G(1) cell extracts, and correlated with a concomitant increase in cyclin A-associated kinase activity. Conversely, a specific antibody neutralizing cyclin A-dependent kinase activity, abolished the capacity of S cell extracts for DNA conversion. We found no evidence for the involvement of cyclin E in the regulation of the conversion reaction. We conclude that cyclin A is necessary for activation of complementary strand synthesis, which we propose as a model reaction to study the cell cycle regulation of the Pol delta-dependent elongation machinery.
Figures
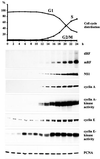
Figure 1
Time course analysis of MVM DNA replication in synchronized A9 cells. Suspension cultures were synchronized in G0/G1 by serum starvation and infected with MVM (MOI = 10 pfu/cell). At 12 h p.i., cells were released from the G1 block by addition of FCS (20% final concentration). Culture samples were taken at 2-h intervals and monitored for cell cycle distribution, production of MVM DNA replication intermediates, and viral and cellular protein expression. Cyclin A- and E-associated kinase activities were determined as described in Materials and Methods. mRF, monomer replicative form DNA; dRF, dimer replicative form DNA.
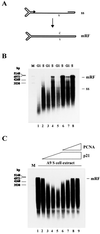
Figure 2
Conversion of MVM ss DNA into RF DNA in extracts from A9 cells synchronized in G1 and S phase. (A) Schematic representation of the conversion reaction. (B) MVM ss DNA (20 ng) was incubated in G1 or S cell extracts corresponding to 40 μg (lanes 1 and 2), 60 μg (lanes 3 and 4), 80 μg (lanes 5 and 6), or 100 μg (lanes 7 and 8) of cellular proteins. (C) MVM ss DNA (20 ng) was incubated in S cell extract alone (lane 1) or supplemented with 0.4 μg (lane 2), 0.8 μg (lane 3), 1.2 μg (lane 4), or 1.6 μg (lanes 5 to 9) of recombinant p21WAF1/CIP1 and additionally 0.4 μg (lane 6), 0.8 μg (lane 7), 1.2 μg (lane 8), or 1.6 μg (lane 9) of recombinant PCNA. Replication products were analyzed by neutral agarose gel electrophoresis (0.8%). ss, single-stranded virion DNA; mRF, monomer replicative form DNA; v, viral strand; c, complementary strand; M, DNA size markers in bp.
Figure 3
Effect of cyclin A on the ability of A9 cell extracts to convert MVM ss DNA into RF. (A) MVM ss DNA (20 ng) was incubated in G1 cell extract alone (80 μg) (lane 1) or in the presence of 0.1 μg (lane 2), 0.2 μg (lane 3), 0.4 μg (lane 4), 0.6 μg (lane 5), or 0.8 μg (lane 6) of GST-cyclin A. Cyclin A-dependent kinase activity was determined by phosphorylation of histone H1 after addition of increasing amounts of GST-cyclin A (0.0 μg, lane 7; 0.1 μg, lane 8; 0.2 μg, lane 9; 0.4 μg, lane 10; 0.8 μg, lane 11; 1.6 μg, lane 12) to G1 cell extract samples (80 μg protein) and cyclin A/cdk complex immunoprecipitation. (B) MVM ss DNA (20 ng) was incubated in S cell extract alone (lane 1) or in the presence of 0.4 μg (lane 2), 0.8 μg (lane 3), 1.2 μg (lane 4), 1.6 μg (lane 5), or 2.0 μg (lanes 6 to 10) of the cyclin A-specific neutralizing mAb E23 and additionally 0.2 μg (lane 7), 0.4 μg (lane 8), 0.8 μg (lane 9), or 1.2 μg (lane 10) of GST-cyclin A. (C) MVM ss DNA (20 ng) was incubated in S cell extract alone (lane 1) or in the presence of 0.4 μg (lane 2), 0.8 μg (lane 3), 1.2 μg (lane 4), 1.6 μg (lane 5), or 2.0 μg (lane 6) of the cyclin A-specific nonneutralizing mAb E72. (D) Samples (80 μg protein) from S cell extract were supplemented with increasing amounts of the cyclin A-specific Abs E23 or E72 (no Ab, lanes 1 and 6; 0.6 μg, lanes 2 and 7; 1.2 μg, lanes 3 and 8; 1.8 μg, lanes 4 and 9), and kinase activities were determined as described in Materials and Methods. mRF, monomer replicative form DNA; M, size markers in bp.
Figure 4
Effect of cyclins A and E on the capacity of A9 G1 cell extracts for conversion of MVM ss DNA into RF. MVM ss DNA (20 ng) was incubated in G1 cell extract (60 μg) alone (lanes 1 and 5) or in the presence of increasing amounts of cyclin A (80 ng, lane 2; 160 ng, lane 3; 320 ng, lane 4) or cyclin E/cdk2 (50 ng, lane 6; 100 ng, lane 7; 200 ng, lane 8). The yields of conversion are shown on the top, whereas corresponding cyclin A- and cyclin E-associated kinase activities are illustrated on the bottom.
Figure 5
Schematic representation of the cellular factors assembled at the MVM virion DNA primer-template junction and thought to be involved in complementary strand elongation. Cyclin A or the cyclin A/cdk2 complex may regulate the conversion reaction by forming part of the elongation machinery and/or phosphorylating some of its constituents.
Similar articles
- In vivo accumulation of cyclin A and cellular replication factors in autonomous parvovirus minute virus of mice-associated replication bodies.
Bashir T, Rommelaere J, Cziepluch C. Bashir T, et al. J Virol. 2001 May;75(9):4394-8. doi: 10.1128/JVI.75.9.4394-4398.2001. J Virol. 2001. PMID: 11287588 Free PMC article. - Cell cycle-dependent dynamic association of cyclin/Cdk complexes with human DNA replication proteins.
Frouin I, Montecucco A, Biamonti G, Hübscher U, Spadari S, Maga G. Frouin I, et al. EMBO J. 2002 May 15;21(10):2485-95. doi: 10.1093/emboj/21.10.2485. EMBO J. 2002. PMID: 12006500 Free PMC article. - NS1- and minute virus of mice-induced cell cycle arrest: involvement of p53 and p21(cip1).
Op De Beeck A, Sobczak-Thepot J, Sirma H, Bourgain F, Brechot C, Caillet-Fauquet P. Op De Beeck A, et al. J Virol. 2001 Nov;75(22):11071-8. doi: 10.1128/JVI.75.22.11071-11078.2001. J Virol. 2001. PMID: 11602746 Free PMC article. - Initiation and termination of DNA replication during S phase in relation to cyclins D1, E and A, p21WAF1, Cdt1 and the p12 subunit of DNA polymerase δ revealed in individual cells by cytometry.
Darzynkiewicz Z, Zhao H, Zhang S, Lee MY, Lee EY, Zhang Z. Darzynkiewicz Z, et al. Oncotarget. 2015 May 20;6(14):11735-50. doi: 10.18632/oncotarget.4149. Oncotarget. 2015. PMID: 26059433 Free PMC article. Review. - Protoparvovirus Interactions with the Cellular DNA Damage Response.
Majumder K, Etingov I, Pintel DJ. Majumder K, et al. Viruses. 2017 Oct 31;9(11):323. doi: 10.3390/v9110323. Viruses. 2017. PMID: 29088070 Free PMC article. Review.
Cited by
- Replication Compartments of Eukaryotic and Bacterial DNA Viruses: Common Themes Between Different Domains of Host Cells.
Knipe DM, Prichard A, Sharma S, Pogliano J. Knipe DM, et al. Annu Rev Virol. 2022 Sep 29;9(1):307-327. doi: 10.1146/annurev-virology-012822-125828. Annu Rev Virol. 2022. PMID: 36173697 Free PMC article. Review. - Efficient parvovirus replication requires CRL4Cdt2-targeted depletion of p21 to prevent its inhibitory interaction with PCNA.
Adeyemi RO, Fuller MS, Pintel DJ. Adeyemi RO, et al. PLoS Pathog. 2014 Apr 3;10(4):e1004055. doi: 10.1371/journal.ppat.1004055. eCollection 2014 Apr. PLoS Pathog. 2014. PMID: 24699724 Free PMC article. - Transforming growth factor-β activates c-Myc to promote palatal growth.
Zhu X, Ozturk F, Liu C, Oakley GG, Nawshad A. Zhu X, et al. J Cell Biochem. 2012 Oct;113(10):3069-85. doi: 10.1002/jcb.24184. J Cell Biochem. 2012. PMID: 22573578 Free PMC article. - High mutant frequency in populations of a DNA virus allows evasion from antibody therapy in an immunodeficient host.
López-Bueno A, Mateu MG, Almendral JM. López-Bueno A, et al. J Virol. 2003 Feb;77(4):2701-8. doi: 10.1128/jvi.77.4.2701-2708.2003. J Virol. 2003. PMID: 12552010 Free PMC article. - Oncolytic rat parvovirus H-1PV, a candidate for the treatment of human lymphoma: In vitro and in vivo studies.
Angelova AL, Aprahamian M, Balboni G, Delecluse HJ, Feederle R, Kiprianova I, Grekova SP, Galabov AS, Witzens-Harig M, Ho AD, Rommelaere J, Raykov Z. Angelova AL, et al. Mol Ther. 2009 Jul;17(7):1164-72. doi: 10.1038/mt.2009.78. Epub 2009 Apr 14. Mol Ther. 2009. PMID: 19367260 Free PMC article.
References
- Reed S I. Annu Rev Cell Biol. 1992;8:529–561. - PubMed
- Nigg E. BioEssays. 1995;17:471–480. - PubMed
- Cardoso M C, Leonhardt H, Nadal-Ginard B. Cell. 1993;74:979–992. - PubMed
- Sobczak-Thepot J, Harper F, Florentin Y, Zindy F, Brechot C, Puvion E. Exp Cell Res. 1993;206:43–48. - PubMed
- Fotedar R, Roberts J M. Cold Spring Harbor Symp Quant Biol. 1991;56:325–333. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources