Phosphorylation of protein kinase N by phosphoinositide-dependent protein kinase-1 mediates insulin signals to the actin cytoskeleton - PubMed (original) (raw)
Phosphorylation of protein kinase N by phosphoinositide-dependent protein kinase-1 mediates insulin signals to the actin cytoskeleton
L Q Dong et al. Proc Natl Acad Sci U S A. 2000.
Abstract
Growth factors such as insulin regulate phosphatidylinositol 3-kinase-dependent actin cytoskeleton rearrangement in many types of cells. However, the mechanism by which the insulin signal is transmitted to the actin cytoskeleton remains largely unknown. Yeast two-hybrid screening revealed that the phosphatidylinositol 3-kinase downstream effector phosphoinositide-dependent protein kinase-1 (PDK1) interacted with protein kinase N (PKN), a Rho-binding Ser/Thr protein kinase potentially implicated in a variety of cellular events, including phosphorylation of cytoskeletal components. PDK1 and PKN interacted in vitro and in intact cells, and this interaction was mediated by the kinase domain of PDK1 and the carboxyl terminus of PKN. In addition to a direct interaction, PDK1 also phosphorylated Thr(774) in the activation loop and activated PKN. Insulin treatment or ectopic expression of the wild-type PDK1 or PKN, but not protein kinase Czeta, induced actin cytoskeleton reorganization and membrane ruffling in 3T3-L1 fibroblasts and Rat1 cells that stably express the insulin receptor (Rat1-IR). However, the insulin-stimulated actin cytoskeleton reorganization in Rat1-IR cells was prevented by expression of kinase-defective PDK1 or PDK1-phosphorylation site-mutated PKN. Thus, phosphorylation by PDK1 appears to be necessary for PKN to transduce signals from the insulin receptor to the actin cytoskeleton.
Figures
Figure 1
Interaction of PKN with PDK1. (A) Schematic representation of PKN constructs used to study PKN and PDK1 interaction and phosphorylation. The consensus PDK1 phosphorylation sequences of several PDK1 substrates are shown. (B) Interaction of PDK1 with PKN in the yeast two-hybrid system. cDNAs containing different regions of PDK1 were amplified by PCR and subcloned into the yeast two-hybrid plasmid pGBT9. The interaction between PDK1 fragments and PKN-CT in SFY526 yeast cells was detected by β-galactosidase filter assays as described (14). +, Positive interaction (blue color) was visualized within 30 min; −, no interaction was detected for 24 h. N, Amino terminus; CD, catalytic domain; PH, pleckstrin homology domain. (C) Interaction between PDK1 and PKN in vitro. Lysates of CHO/IR/PDK1 cells (11) were incubated with GST/AF3-PKNK644E (lane 1) or GST/PKN(ΔC) (lane 2) bound to glutathione-Sepharose beads (Sigma). The bound proteins were resolved by SDS/PAGE, transferred onto a nitrocellulose membrane, and examined by immunoblotting with antibody to the hemagglutinin tag (Top). Five percent of cell lysates used in the immunoprecipitation were loaded as a control (Middle). The GST fusion proteins used in the experiments were separated by SDS/PAGE and visualized by Coomassie blue staining (Bottom). Results are representative of three independent experiments. (D) Interaction of PDK1 and PKN in intact cells. CHO/IR cells were serum-starved overnight and treated with or without wortmannin (WT, 200 nM) or LY294002 (LY, 200 μM) for 1 h. Cells then were treated with (+) or without (−) 10 nM insulin for 10 min. Cell lysates were incubated for 8 h at 4°C with affinity-purified polyclonal antibody to PDK1 (11) or NIg bound to protein A beads. PKN coimmunoprecipitated with PDK1 was detected with a mAb to PKN (Transduction Laboratories).
Figure 2
Phosphorylation and activation of PKN by PDK1. (A) Time course study of PKN phosphorylation. The FLAG-tagged full-length PKN was immunoprecipitated from CHO/IR/PKN cells with the M2 anti-FLAG antibody. In vitro phosphorylation was carried out in the presence (+) or absence (−) of immunoaffinity-purified PDK1 (11). Reactions were stopped at indicated times. The phosphorylated proteins were eluted from beads by boiling in SDS-sample buffer, separated by SDS/PAGE, transferred to a nitrocellulose membrane, and visualized by autoradiography (Upper). The same membrane was reblotted with anti-FLAG antibody to ensure equal loading of proteins in each lane (Lower). (B) Phosphorylation of AF3-PKN by PDK1. AF3-PKN (15) was transiently expressed in CHO/IR cells, immunoprecipitated by anti-FLAG antibody, and phosphorylated in vitro in the presence of immunoaffinity-purified PDK1 or PDK1K114G, according to a protocol similar to that described previously (11). The phosphorylation of AF3-PKN was visualized by autoradiography. (C) Phosphoamino acid analysis of AF3-PKN. The 32P-labeled wild-type (WT) or T774A mutant of AF3-PKN bands were excised from the membrane, hydrolyzed, and analyzed by two-dimensional thin-layer electrophoresis, according to a protocol described previously (16). (D) The effect of PDK1 on PKN phosphorylation in cells. CHO/IR cells were transfected with PKN and PDK1, _in vivo-_labeled with 32P-orthophosphate, and treated with (+) or without (−) insulin as described in Materials and Methods. Data are mean ± SEM from four independent experiments. Background values obtained from NIg precipitates were subtracted from all values. (E) Overexpression of PDK1 stimulates PKN phosphorylation at Thr774 in intact cells. CHO/IR cells were cotransfected with FLAG-tagged PKN or PKNT774A and either myc-tagged PDK1 or PDK1K114G as indicated. Cell lysates were immunoprecipitated with antibody to the FLAG tag. One-eighth of the immunoprecipitates were separated by SDS/PAGE and analyzed by Western blot with antibody specific to phospho-Thr816 of PRK2 (Top). Two-thirds of the immunoprecipitates were used for Western blot with antibody to the FLAG tag (Middle). Expression of PDK1 in these cells was detected by Western blot of cell lysate (1/20 of total) using antibody to the myc tag (Bottom). (F) Insulin stimulates the phosphorylation of endogenous PKN at Thr774 in CHO/IR cells. CHO/IR cells were serum-starved overnight and treated with or without wortmannin (WT, 200 nM) or LY294002 (LY, 200 μM) for 1 h. Cells then were treated with (+) or without (−) 10 nM insulin for 10 min. Cell lysates were incubated for 8 h at 4°C with mAb to PKN (Transduction Laboratories) or NIg bound to protein G beads. One-eighth of the immunoprecipitates were separated by SDS/PAGE and analyzed by Western blot with antibody specific to phospho-Thr816 of PRK2 (Upper). Two-thirds of the immunoprecipitates were used for Western blot with a mAb to PKN (Transduction Laboratories) (Lower).
Figure 3
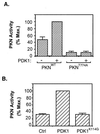
Activation of PKN by PDK1. (A) Activation of PKN by PDK1 in vitro. The FLAG-tagged wild-type PKN or PKNT774A was immunoprecipitated from serum-starved CHO/IR cells transiently expressing these proteins, phosphorylated in vitro in the presence (+) or absence (−) of affinity-purified PDK1, and assayed for phosphorylation of the S6 peptide. Data are representative of three independent experiments, with bars representing means of duplicate determinations. (B) Activation of PKN by PDK1 in cells. CHO/IR cells were cotransfected with cDNAs encoding PKN, pcDNA/PDK1, or pcDNA/PDK1K114G and the control vector pcDNA. Twenty-four hours after transfection, cells were lysed and activity of the immunoprecipitated PKN was assayed as described in Materials and Methods. Data are mean ± SD of two independent experiments with duplicate determinations.
Figure 4
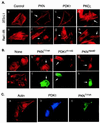
Effects of PDK1 and PKN on actin cytoskeleton reorganization. (A) Ectopic expression of PDK1 and PKN induces actin cytoskeleton reorganization. 3T3-L1 (Upper) or Rat1-IR (Lower) cells were transfected with plasmids encoding FLAG-PKN, myc-PDK1, or myc-PKCζ, respectively. Arrows indicate cells expressing the recombinant proteins as determined by staining with antibodies to the tags. Actin cytoskeleton was detected by a rhodamine-phalloidin stain. (B) Ectopic expression of mutant PKN or PDK1 blocks insulin-induced actin cytoskeleton reorganization. Rat1-IR cells (a and e) or cells transiently expressing FLAG-PKNT774A (b and f), myc-PDK1K114G (c and g), or FLAG-PKNK644D (d and h) were serum starved for 4 h and treated with 10 nM insulin for 10 min (b_–_h). Actin cytoskeleton was detected by a rhodamine-phalloidin stain (a_–_e). The expression of the recombinant proteins in cells was determined by staining with the anti-FLAG or anti-myc antibodies, followed by Alexa Fluor 488-conjugated goat anti-mouse Ig (f_–_h). Images are representative of those in three independent experiments. (C) Overexpression of PKNT774A inhibits PDK1-induced actin stress fiber reorganization. Rat1-IR cells were cotransfected with FLAG-tagged PKNT774A and myc-tagged PDK1. The expression of the recombinant proteins in cells was determined by staining with the anti-FLAG or anti-myc antibodies, followed by Alexa Fluor 488-conjugated goat anti-mouse Ig (green, for PKNT774A) or Alexa Fluor 350-conjugated goat anti-rabbit Ig (blue, for PDK1), respectively. Actin cytoskeleton was detected by a rhodamine-phalloidin stain (red). Results are representative of two independent experiments in which the inhibition of PDK1-induced actin stress fiber reorganization was observed in approximately 65% of cells coexpressing PKNT774A.
Similar articles
- Mechanism of phosphorylation of protein kinase B/Akt by a constitutively active 3-phosphoinositide-dependent protein kinase-1.
Wick MJ, Dong LQ, Riojas RA, Ramos FJ, Liu F. Wick MJ, et al. J Biol Chem. 2000 Dec 22;275(51):40400-6. doi: 10.1074/jbc.M003937200. J Biol Chem. 2000. PMID: 11006271 - 3-Phosphoinositide-dependent protein kinase 1 (PDK1) phosphorylates and activates the p70 S6 kinase in vivo and in vitro.
Alessi DR, Kozlowski MT, Weng QP, Morrice N, Avruch J. Alessi DR, et al. Curr Biol. 1998 Jan 15;8(2):69-81. doi: 10.1016/s0960-9822(98)70037-5. Curr Biol. 1998. PMID: 9427642 - Role of the actin cytoskeleton in insulin action.
Tsakiridis T, Tong P, Matthews B, Tsiani E, Bilan PJ, Klip A, Downey GP. Tsakiridis T, et al. Microsc Res Tech. 1999 Oct 15;47(2):79-92. doi: 10.1002/(SICI)1097-0029(19991015)47:2<79::AID-JEMT1>3.0.CO;2-S. Microsc Res Tech. 1999. PMID: 10523787 Review. - PDK2: the missing piece in the receptor tyrosine kinase signaling pathway puzzle.
Dong LQ, Liu F. Dong LQ, et al. Am J Physiol Endocrinol Metab. 2005 Aug;289(2):E187-96. doi: 10.1152/ajpendo.00011.2005. Am J Physiol Endocrinol Metab. 2005. PMID: 16014356 Review.
Cited by
- Novel roles of PRK1 and PRK2 in cilia and cancer biology.
Patel H, Li J, Herrero A, Kroboth J, Byron A, Kriegsheim AV, Brunton V, Carragher N, Hurd T, Frame M. Patel H, et al. Sci Rep. 2020 Mar 3;10(1):3902. doi: 10.1038/s41598-020-60604-3. Sci Rep. 2020. PMID: 32127582 Free PMC article. - The human papillomavirus type 16 E6 oncoprotein activates mTORC1 signaling and increases protein synthesis.
Spangle JM, Münger K. Spangle JM, et al. J Virol. 2010 Sep;84(18):9398-407. doi: 10.1128/JVI.00974-10. Epub 2010 Jul 14. J Virol. 2010. PMID: 20631133 Free PMC article. - 1H, 15N and 13C resonance assignments of the HR1c domain of PRK1, a protein kinase C-related kinase.
Sophocleous G, Wood G, Owen D, Mott HR. Sophocleous G, et al. Biomol NMR Assign. 2020 Oct;14(2):245-250. doi: 10.1007/s12104-020-09954-7. Epub 2020 Jun 4. Biomol NMR Assign. 2020. PMID: 32500230 Free PMC article. - Steady-state kinetic mechanism of PDK1.
Gao X, Harris TK. Gao X, et al. J Biol Chem. 2006 Aug 4;281(31):21670-21681. doi: 10.1074/jbc.M602448200. Epub 2006 May 31. J Biol Chem. 2006. PMID: 16737971 Free PMC article. - PAK and other Rho-associated kinases--effectors with surprisingly diverse mechanisms of regulation.
Zhao ZS, Manser E. Zhao ZS, et al. Biochem J. 2005 Mar 1;386(Pt 2):201-14. doi: 10.1042/BJ20041638. Biochem J. 2005. PMID: 15548136 Free PMC article. Review.
References
- Zigmond S H. Curr Opin Cell Biol. 1996;8:66–73. - PubMed
- Anand-Apte B, Zetter B. Stem Cells. 1997;15:259–267. - PubMed
- Schmidt A, Hall M N. Annu Rev Cell Dev Biol. 1998;14:305–338. - PubMed
- Wennstrom S, Siegbahn A, Yokote K, Arvidsson A K, Heldin C H, Mori S, Claesson W L. Oncogene. 1994;9:651–660. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous