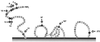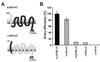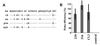Simian immunodeficiency virus utilizes human and sooty mangabey but not rhesus macaque STRL33 for efficient entry - PubMed (original) (raw)
Simian immunodeficiency virus utilizes human and sooty mangabey but not rhesus macaque STRL33 for efficient entry
S Pöhlmann et al. J Virol. 2000 Jun.
Abstract
It has been established that many simian immunodeficiency virus (SIV) isolates utilize the orphan receptors GPR15 and STRL33 about as efficiently as the chemokine receptor CCR5 for entry into target cells. Most studies were performed, however, with coreceptors of human origin. We found that SIV from captive rhesus macaques (SIVmac) can utilize both human and simian CCR5 and GPR15 with comparable efficiencies. Strikingly, however, only human STRL33 (huSTRL33), not rhesus macaque STRL33 (rhSTRL33), functioned efficiently as an entry cofactor for a variety of isolates of SIVmac and SIV from sooty mangabeys. A single amino acid substitution of S30R in huSTRL33 impaired coreceptor activity, and the reverse change in rhSTRL33 greatly increased coreceptor activity. In comparison, species-specific sequence variations in N-terminal tyrosines in STRL33 had only moderate effects on SIV entry. These results show that a serine residue located just outside of the cellular membrane in the N terminus of STRL33 is critical for SIV coreceptor function. Interestingly, STRL33 derived from sooty mangabeys, a natural host of SIV, also contained a serine at the corresponding position and was used efficiently as an entry cofactor. These results suggest that STRL33 is not a relevant coreceptor in the SIV/macaque model but may play a role in SIV replication and transmission in naturally infected sooty mangabeys.
Figures
FIG. 1
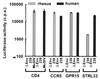
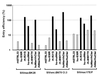
SIVmac239 does not use rhSTRL33 for efficient entry. 293T cells were cotransfected with plasmids expressing human or macaque CD4 and the indicated entry cofactors of human and rhesus macaque origin. At 1 day posttransfection the cells were detached from the plates, seeded in 48-well dishes, and infected in triplicate with intact luciferase reporter viruses containing 50 ng of p27 antigen. Luciferase activities in the cellular extracts were measured at 3 days postinfection. Error bars give standard deviations from average values measured in three independent infections. Similar results were obtained in three additional experiments. As controls the transfected cells were infected with envelope-deleted SIVmac239 (Δenv) or with a chimeric SIV containing the murine leukemia virus glycoprotein (MuSIV) (63). Luciferase values obtained with rhSTRL33 (1,885 ± 61) were about 13-fold lower than those obtained with huSTRL33 (24,043 ± 2,993).
FIG. 2
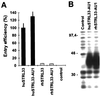
Different SIV entry efficiencies are not due to inefficient expression of rhSTRL33. (A) 293T cells were cotransfected with CD4 and expression constructs for wild-type and AU1-tagged huSTRL33 and rhSTRL33. SIVmac239 entry was determined as described in the legend to Fig. 1. The results represent average values of three independent infections. (B) 293T cells were transfected with vectors expressing AU1-tagged huSTRL33 or rhSTRL33. Protein expression was verified by Western blot analysis as described in Materials and Methods. Control, cell extracts derived from mock-transfected 293T cells.
FIG. 3
Schematic presentation of the N-terminal region and the extracellular domains of rhSTRL33. Amino acid variations in huSTRL33 compared to the rhSTRL33 sequence are indicated. huSTRL33 and rhSTRL33 are 94% identical at the amino acid level, with most differences clustered at the amino terminus (20, 46). Differences between huSTRL33 and rhSTRL33 are indicated by the shaded residues. The numbers give the amino acid positions in rhSTRL33. Bar, cell membrane.
FIG. 4
Species-specific sequence variations in the N terminus of STRL33 determine SIV coreceptor function. (A) Schematic presentation of the AU1-tagged STRL33 recombinants with an huSTRL33 N terminus and a rhSTRL33 C terminus (huN/rhC) and with an rhSTRL33 N terminus and an huSTRL33 C terminus (rhN/huC). (B) 293T cells were cotransfected with CD4 and expression constructs for the indicated STRL33 variants. SIVmac239 entry was tested as described in the legend to Fig. 1. The results represent average values of three independent infections. Control, cell extracts derived from mock-transfected 293T cells.
FIG. 5
Amino acid substitution of S31R impairs functional activity of rhSTRL33 as an SIVmac239 entry cofactor. Mutational analysis of rhSTRL33 and huSTRL33. The specific mutations compared to the original rhSTRL33 and huSTRL33 amino acid sequences are shown at the left. Dashes indicate amino acid identity, and dots indicate gaps. The relative entry efficiencies compared to human STRL33 are shown at the right. Average values from three infections are shown, and comparable results were obtained with independent virus stocks.
FIG. 6
Serine 31 is critical for STRL33 coreceptor function of several SIV isolates. SIVmacBK28, SIVsmΔB670 cl.3, and SIVmac17E/F Env-pseudotyped GFP reporter viruses were used to infect cells coexpressing CD4 and the indicated STRL33 variants. Infection efficiency (percentage of GFP-positive cells) is shown relative to that obtained for huSTRL33 (100%). Comparable results were obtained in two independent experiments.
FIG. 7
smSTRL33 mediates efficient SIV entry. (A) Sequence variations at the N terminus of STRL33 derived from humans (hu), rhesus macaques (rh), sooty mangabeys (sm), pig tail macaques (ptm), and African green monkeys (agm) (20, 46). Dashes, identity with the human-derived STRL33 sequence; dots, gaps introduced to optimize the alignment. The position of the serine residue, which is critical for STRL33 coreceptor function, is boxed. (B) Entry of luciferase reporter viruses pseudotyped with SIVmac239, SIVmac316, and SIVsmΔB670 cl.3 Env into cells coexpressing CD4 and smSTRL33. Infections were performed as indicated in Materials and Methods. Entry efficiency is shown relative to that obtained for huSTRL33. Data represent average values obtained for three independent infections. Control, cell extracts derived from mock-transfected 293T cells infected with pseudotyped luciferase reporter viruses.
Similar articles
- Use of GPR1, GPR15, and STRL33 as coreceptors by diverse human immunodeficiency virus type 1 and simian immunodeficiency virus envelope proteins.
Edinger AL, Hoffman TL, Sharron M, Lee B, O'Dowd B, Doms RW. Edinger AL, et al. Virology. 1998 Sep 30;249(2):367-78. doi: 10.1006/viro.1998.9306. Virology. 1998. PMID: 9791028 - Simian immunodeficiency viruses of diverse origin can use CXCR4 as a coreceptor for entry into human cells.
Owen SM, Masciotra S, Novembre F, Yee J, Switzer WM, Ostyula M, Lal RB. Owen SM, et al. J Virol. 2000 Jun;74(12):5702-8. doi: 10.1128/jvi.74.12.5702-5708.2000. J Virol. 2000. PMID: 10823878 Free PMC article. - The function of simian chemokine receptors in the replication of SIV.
Marx PA, Chen Z. Marx PA, et al. Semin Immunol. 1998 Jun;10(3):215-23. doi: 10.1006/smim.1998.0135. Semin Immunol. 1998. PMID: 9653048 Review. - SIV Coreceptor Specificity in Natural and Non-Natural Host Infection: Implications for Cell Targeting and Differential Outcomes from Infection.
Wetzel KS, Elliott STC, Collman RG. Wetzel KS, et al. Curr HIV Res. 2018;16(1):41-51. doi: 10.2174/1570162X15666171124121805. Curr HIV Res. 2018. PMID: 29173179 Review.
Cited by
- Cloning and analysis of sooty mangabey alternative coreceptors that support simian immunodeficiency virus SIVsmm entry independently of CCR5.
Elliott ST, Riddick NE, Francella N, Paiardini M, Vanderford TH, Li B, Apetrei C, Sodora DL, Derdeyn CA, Silvestri G, Collman RG. Elliott ST, et al. J Virol. 2012 Jan;86(2):898-908. doi: 10.1128/JVI.06415-11. Epub 2011 Nov 16. J Virol. 2012. PMID: 22090107 Free PMC article. - Simian Immunodeficiency Virus SIVagm Efficiently Utilizes Non-CCR5 Entry Pathways in African Green Monkey Lymphocytes: Potential Role for GPR15 and CXCR6 as Viral Coreceptors.
Riddick NE, Wu F, Matsuda K, Whitted S, Ourmanov I, Goldstein S, Goeken RM, Plishka RJ, Buckler-White A, Brenchley JM, Hirsch VM. Riddick NE, et al. J Virol. 2015 Dec 9;90(5):2316-31. doi: 10.1128/JVI.02529-15. J Virol. 2015. PMID: 26656714 Free PMC article. - Genetic identity and biological phenotype of a transmitted/founder virus representative of nonpathogenic simian immunodeficiency virus infection in African green monkeys.
Gnanadurai CW, Pandrea I, Parrish NF, Kraus MH, Learn GH, Salazar MG, Sauermann U, Töpfer K, Gautam R, Münch J, Stahl-Hennig C, Apetrei C, Hahn BH, Kirchhoff F. Gnanadurai CW, et al. J Virol. 2010 Dec;84(23):12245-54. doi: 10.1128/JVI.01603-10. Epub 2010 Sep 29. J Virol. 2010. PMID: 20881048 Free PMC article. - Effect of semen and seminal amyloid on vaginal transmission of simian immunodeficiency virus.
Münch J, Sauermann U, Yolamanova M, Raue K, Stahl-Hennig C, Kirchhoff F. Münch J, et al. Retrovirology. 2013 Dec 5;10:148. doi: 10.1186/1742-4690-10-148. Retrovirology. 2013. PMID: 24308721 Free PMC article. - TCR triggering transcriptionally downregulates CCR5 expression on rhesus macaque CD4(+) T-cells with no measurable effect on susceptibility to SIV infection.
Minang JT, Trivett MT, Barsov EV, Del Prete GQ, Trubey CM, Thomas JA, Gorelick RJ, Piatak M Jr, Ott DE, Ohlen C. Minang JT, et al. Virology. 2011 Jan 5;409(1):132-40. doi: 10.1016/j.virol.2010.10.005. Epub 2010 Oct 28. Virology. 2011. PMID: 21035160 Free PMC article.
References
- Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. CC-CKR5: a RANTES, MIP-1alpha, MIP-1beta receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. - PubMed
- Alkhatib G, Liao F, Berger E A, Farber J M, Peden K W. A new SIV co-receptor, STRL33. Nature. 1997;388:238. - PubMed
- Anderson M G, Hauer D, Sharma D P, Joag S V, Narayan O, Zink M C, Clements J E. Analysis of envelope changes acquired by SIVmac239 during neuroadaption in rhesus macaques. Virology. 1993;195:616–626. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases