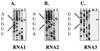Characterization and construction of functional cDNA clones of Pariacoto virus, the first Alphanodavirus isolated outside Australasia - PubMed (original) (raw)
Characterization and construction of functional cDNA clones of Pariacoto virus, the first Alphanodavirus isolated outside Australasia
K N Johnson et al. J Virol. 2000 Jun.
Abstract
Pariacoto virus (PaV) was recently isolated in Peru from the Southern armyworm (Spodoptera eridania). PaV particles are isometric, nonenveloped, and about 30 nm in diameter. The virus has a bipartite RNA genome and a single major capsid protein with a molecular mass of 39.0 kDa, features that support its classification as a Nodavirus. As such, PaV is the first Alphanodavirus to have been isolated from outside Australasia. Here we report that PaV replicates in wax moth larvae and that PaV genomic RNAs replicate when transfected into cultured baby hamster kidney cells. The complete nucleotide sequences of both segments of the bipartite RNA genome were determined. The larger genome segment, RNA1, is 3,011 nucleotides long and contains a 973-amino-acid open reading frame (ORF) encoding protein A, the viral contribution to the RNA replicase. During replication, a 414-nucleotide long subgenomic RNA (RNA3) is synthesized which is coterminal with the 3' end of RNA1. RNA3 contains a small ORF which could encode a protein of 90 amino acids similar to the B2 protein of other alphanodaviruses. RNA2 contains 1,311 nucleotides and encodes the 401 amino acids of the capsid protein precursor alpha. The amino acid sequences of the PaV capsid protein and the replicase subunit share 41 and 26% identity with homologous proteins of Flock house virus, the best characterized of the alphanodaviruses. These and other sequence comparisons indicate that PaV is evolutionarily the most distant of the alphanodaviruses described to date, consistent with its novel geographic origin. Although the PaV capsid precursor is cleaved into the two mature capsid proteins beta and gamma, the amino acid sequence at the cleavage site, which is Asn/Ala in all other alphanodaviruses, is Asn/Ser in PaV. To facilitate the investigation of PaV replication in cultured cells, we constructed plasmids that transcribed full-length PaV RNAs with authentic 5' and 3' termini. Transcription of these plasmids in cells recreated the replication of PaV RNA1 and RNA2, synthesis of subgenomic RNA3, and translation of viral proteins A and alpha.
Figures
FIG. 1
Schematic representation of the transcription plasmid TVT7R(0,0). (A) DNA sequence of the region encompassing the T7 promoter (underlined), transcription start site, and site of RNA cleavage (both indicated with arrowheads). The _Bbs_I recognition sequences are shown in italics, and the nucleotides that remain following excision of the short stuffer fragment by _Bbs_I digestion are shown in bold. Positions of the HDV antigenomic ribozyme (Rz) and T7 terminator sequences are shown. (B) Positioning of PaV cDNAs in the transcription plasmid.
FIG. 2
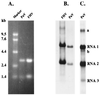
PaV RNAs and replication in BHK-21 cells. (A) RNAs extracted from purified PaV and FHV virions were resolved by electrophoresis in a 1% agarose-formaldehyde gel along with RNA markers (Gibco/BRL) and visualized by ethidium bromide staining. Sizes of the RNA markers are indicated on the left. (B) BHK-21 cells were transfected with 1 μg of PaV virion RNA or with 0.5 μg of FHV virion RNA and incubated at 28°C. After 22 h of incubation, actinomycin D was added at 5 μg/ml; 30 min later, replicating RNAs were metabolically labeled by incorporation of [3H]uridine for a period of 2 h before total cellular RNA was harvested. RNAs were resolved by electrophoresis on a 1% agarose-formaldehyde gel and visualized by fluorography. (C) The PaV lane of the autoradiogram in panel B was overexposed so that PaV RNA3 could be visualized. PaV RNA1, RNA2, and RNA3 are identified on the right, as are two additional minor RNAs, bands a and b.
FIG. 3
Sequences of the 5′ and 3′ termini of PaV RNA1 and RNA2. The consensus sequences of the termini of PaV RNA1 and RNA2 were compiled using three independent methods, and the sequences generated by each method are shown. The homodimer junction sequences have been separated into their corresponding 5′ and 3′ termini for clarity of presentation. In each case, the number of clones (x) having the particular sequence out of the total number of clones examined (y) is expressed as x/y in the far right column. The consensus terminal sequences derived from the tabulated data are shown in bold for each RNA. The termini of the primer extension products corresponded to the nucleotides indicated by the arrowheads. We attribute the larger products to extension on capped RNAs.
FIG. 4
Analysis of the 5′ termini of genomic RNA1 and RNA2 and subgenomic RNA3 by primer extension. (A and B) RNAs extracted from PaV virions were used as templates for extension of primers designed to anneal to nt 76 to 98 of RNA1 (A, lane P) and 98 to 119 of RNA2 (B, lane P). Dideoxynucleotide sequencing ladders of plasmids containing 5′ RACE products of RNA1 and RNA2 were generated using the same two primers and are shown for reference. (C) BHK-21 cells were transfected with 1 μg of PaV virion RNA (lane P) or mock transfected (lane M) and incubated at 28°C. After 24 h of incubation, total cellular RNA was harvested and used as a template for extension of a primer designed to anneal to nt 2692 to 2712 of RNA1. A dideoxynucleotide sequencing ladder generated using the same primer and plasmid pPaV1(0,0) is shown for reference. For simplicity, in all panels the individual lanes of the sequencing ladders are labeled with the complement of the terminating dideoxynucleotide.
FIG. 5
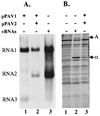
Replication of RNAs and synthesis of viral proteins in cells transfected with PaV cDNA clones. (A) BHK-21 cells were infected with vTF7-3 at an MOI of 10 PFU/cell. One hour postinfection, the cells were transfected with 5 μg of pPaV1(0,0) (lane 1), 2.5 μg each of pPaV1(0,0) and pPaV2(0,0) (lane 2) or 1 μg of PaV virion RNA (lane 3) and incubated at 28°C. After 22 h of incubation, actinomycin D was added at 5 μg/ml; 30 min later, replicating RNAs were metabolically labeled by incorporation of [3H]uridine (20 μCi/ml) for 4 h before total cellular RNA was harvested. RNAs were resolved by electrophoresis on a 1% agarose-formaldehyde gel and visualized by fluorography. PaV RNA1, RNA2, and RNA3 are identified on the left. (B) BHK-21 cells were infected with vTF7-3 at an MOI of 10 PFU/cell. One hour postinfection, the cells were transfected with water (lane 1), 1 μg of PaV virion RNA (lane 2), or 2.5 μg each of pPaV1(0,0) and pPaV2(0,0) (lane 3) and incubated at 28°C. After 44 h of incubation, actinomycin D was added at 5 μg/ml and incubation continued. Following a 0.5-h preincubation in methionine-cysteine-free medium, 48 h posttransfection proteins were labeled with [35S]methionine-cysteine for a period of 2 h. Cytoplasmic extracts were harvested and resolved by SDS-PAGE on a 12.5% gel, and the labeled proteins were visualized by autoradiography. PaV proteins A and α are identified on the right.
FIG. 6
Schematic representation of the arrangement of the ORFs encoded by the PaV genomic RNA1 and RNA2. The horizontal lines represent the RNAs, with vertical lines above and below the RNA indicating positions of methionine codons and termination codons, respectively. For each RNA, the three frames on the positive-sense RNA are shown. The 5′ end of the subgenomic RNA3 is indicated by the dotted vertical line. Bold arrows indicate the four major ORFs which are predicted to encode protein A (RdRp catalytic subunit), protein B1, protein B2, and capsid precursor protein α (1 to 4, respectively). The scale indicates length of RNAs in nucleotides.
FIG. 7
Alignment of amino acid sequences of capsid protein precursors α from all available alphanodaviruses, generated using the GCG program PILEUP. The number of amino acids from the N terminus of protein α is shown at the left. Amino acids that are conserved in three or more of the viruses are shaded. The site of cleavage of the capsid protein precursor α into the two mature capsid proteins β and γ is indicated with an arrow below the alignment. The conserved catalytic Asp residue (75 for FHV) is indicated with an asterisk. Above the alignment, the secondary structural elements as determined from the crystal structure of BBV and labeled according to Johnson and Reddy (25) are indicated as follows: arrows, regions of β sheet; solid lines, regions of α helix; dotted lines; other regions of peptide that are visible in the BBV crystal structure.
FIG. 8
Genotypic relationships among the capsid protein precursors of five alphanodaviruses. (A) Phenogram produced based on the distance matrix generated from the alignment shown in Fig. 7; (B) percent amino acid identity among protein α of the five viruses.
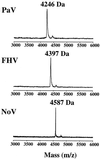
FIG. 9
MALDI-TOF spectrum of protein γ. Whole virus particles were analyzed by MALDI-TOF mass spectrometry. The part of the spectrum which contains protein γ is shown for three nodaviruses: PaV, FHV, and NoV. In each case, the molecular mass of the γ peak is indicated.
Similar articles
- Recovery of infectious pariacoto virus from cDNA clones and identification of susceptible cell lines.
Johnson KN, Ball LA. Johnson KN, et al. J Virol. 2001 Dec;75(24):12220-7. doi: 10.1128/JVI.75.24.12220-12227.2001. J Virol. 2001. PMID: 11711613 Free PMC article. - Recovery of infectivity from cDNA clones of nodamura virus and identification of small nonstructural proteins.
Johnson KL, Price BD, Ball LA. Johnson KL, et al. Virology. 2003 Jan 20;305(2):436-51. doi: 10.1006/viro.2002.1769. Virology. 2003. PMID: 12573589 - Flock house virus: down-regulation of subgenomic RNA3 synthesis does not involve coat protein and is targeted to synthesis of its positive strand.
Zhong W, Rueckert RR. Zhong W, et al. J Virol. 1993 May;67(5):2716-22. doi: 10.1128/JVI.67.5.2716-2722.1993. J Virol. 1993. PMID: 8474170 Free PMC article. - Functional analysis of structural motifs in dicistroviruses.
Nakashima N, Uchiumi T. Nakashima N, et al. Virus Res. 2009 Feb;139(2):137-47. doi: 10.1016/j.virusres.2008.06.006. Epub 2008 Jul 25. Virus Res. 2009. PMID: 18621089 Review. - The insect reservoir of biodiversity for viruses and for antiviral mechanisms.
Olmo RP, Martins NE, Aguiar ERGR, Marques JT, Imler JL. Olmo RP, et al. An Acad Bras Cienc. 2019;91 Suppl 3:e20190122. doi: 10.1590/0001-3765201920190122. Epub 2019 Jun 3. An Acad Bras Cienc. 2019. PMID: 31166476 Review.
Cited by
- Characterization of Nervous Necrosis Virus (NNV) Nonstructural Protein B2 and Its Enhancement on Virus Proliferation.
Zhang Y, Dong F, Xing J, Tang X, Sheng X, Chi H, Zhan W. Zhang Y, et al. Viruses. 2022 Dec 17;14(12):2818. doi: 10.3390/v14122818. Viruses. 2022. PMID: 36560822 Free PMC article. - Betanodavirus induces oxidative stress-mediated cell death that prevented by anti-oxidants and zfcatalase in fish cells.
Chang CW, Su YC, Her GM, Ken CF, Hong JR. Chang CW, et al. PLoS One. 2011;6(10):e25853. doi: 10.1371/journal.pone.0025853. Epub 2011 Oct 3. PLoS One. 2011. PMID: 21991373 Free PMC article. - Internal initiation is responsible for synthesis of Wuhan nodavirus subgenomic RNA.
Qiu Y, Cai D, Qi N, Wang Z, Zhou X, Zhang J, Hu Y. Qiu Y, et al. J Virol. 2011 May;85(9):4440-51. doi: 10.1128/JVI.02410-10. Epub 2011 Feb 16. J Virol. 2011. PMID: 21325414 Free PMC article. - Newcastle Disease Virus V Protein Targets Phosphorylated STAT1 to Block IFN-I Signaling.
Qiu X, Fu Q, Meng C, Yu S, Zhan Y, Dong L, Song C, Sun Y, Tan L, Hu S, Wang X, Liu X, Peng D, Liu X, Ding C. Qiu X, et al. PLoS One. 2016 Feb 9;11(2):e0148560. doi: 10.1371/journal.pone.0148560. eCollection 2016. PLoS One. 2016. PMID: 26859759 Free PMC article. - Development of reverse genetics systems and investigation of host response antagonism and reassortment potential for Cache Valley and Kairi viruses, two emerging orthobunyaviruses of the Americas.
Dunlop JI, Szemiel AM, Navarro A, Wilkie GS, Tong L, Modha S, Mair D, Sreenu VB, Da Silva Filipe A, Li P, Huang YS, Brennan B, Hughes J, Vanlandingham DL, Higgs S, Elliott RM, Kohl A. Dunlop JI, et al. PLoS Negl Trop Dis. 2018 Oct 29;12(10):e0006884. doi: 10.1371/journal.pntd.0006884. eCollection 2018 Oct. PLoS Negl Trop Dis. 2018. PMID: 30372452 Free PMC article.
References
- Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- S10 RR011329/RR/NCRR NIH HHS/United States
- P30 CA013148/CA/NCI NIH HHS/United States
- P30 CA13148-27/CA/NCI NIH HHS/United States
- AI18270/AI/NIAID NIH HHS/United States
- R01 AI018270/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials