The Autographa californica nuclear polyhedrosis virus p143 gene encodes a DNA helicase - PubMed (original) (raw)
The Autographa californica nuclear polyhedrosis virus p143 gene encodes a DNA helicase
V V McDougal et al. J Virol. 2000 Jun.
Abstract
The P143 protein of Autographa californica nuclear polyhedrosis virus is essential for replication of viral DNA. To determine the function of P143, the protein was purified to near homogeneity from recombinant baculovirus-infected cells that overexpress P143. ATPase activity copurified with P143 protein during purification and also during gel filtration at a high salt concentration. The ATPase activity did not require the presence of single-stranded DNA, but was stimulated fourfold by the addition of single-stranded DNA. The ATPase activity of P143 had a K(m) of 60 microM and a turnover of 4.5 molecules of ATP hydrolyzed/s/molecule of enzyme, indicating moderate affinity for ATP and high catalytic efficiency. P143 unwound a 40-nucleotide primer in an ATP-dependent manner, indicating that the enzyme possesses in vitro DNA helicase activity. Based on this result, it seems likely that P143 functions as a helicase in viral DNA replication.
Figures
FIG. 1
Purification of P143. Nuclear extracts (NE) from vAc-P143 infected Sf9 cells (lane 3) were precipitated with Polymin-P, the pellet was resuspended in high-salt buffer to release proteins, and the supernatant was precipitated by ammonium sulfate. The resuspended pellet (lane 4) was subjected to chromatography on heparin. Peak fractions from the heparin column were pooled (lane 5) and fractionated on a Mono S column. Mono S peak fractions (lane 6) were diluted and loaded onto an ssDNA agarose. Lane 7, 4 μg of protein of the peak fraction from ssDNA agarose; lane 2, crude nuclear extract prepared from cells infected with RP6-S/C, the parental virus; lane 1, positions of protein molecular markers are (in kilodaltons) on the left. The arrow on the right indicates the position of P143. Samples were separated on an SDS–8% polyacrylamide gel and stained with Coomassie blue.
FIG. 2
Binding of purified P143 to dsDNA and ssDNA. Lanes 1 to 8, ssDNA probe; lanes 9 to 15, dsDNA probe. The molar ratio of purified P143 to DNA is indicated at the top. Reaction mixtures were separated on a native 3.5% acrylamide–TBE gel, dried, and exposed to film.
FIG. 3
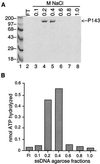
Test of ssDNA agarose column fractions for ATPase activity. (A) Purification of P143 by affinity chromatography on ssDNA agarose. Peak fractions from the Mono S column were loaded onto an ssDNA agarose gravity column and eluted with an NaCl step gradient. Proteins in each fraction were separated on an SDS–8% polyacrylamide gel and stained with Coomassie blue. Lanes 4 and 5 contain P143 purified to near homogeneity. The positions of protein molecular size markers in lane 1 are shown (in kilodaltons) on the left. The arrow on the right indicates the position of P143. (B) ATPase activity. Each fraction was separately dialyzed to 50 mM KCl, and 1 μl of each was assayed for ATPase activity.
FIG. 4
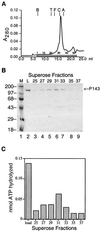
Gel filtration of P143. (A) P143 peak fractions from ssDNA agarose chromatography were adjusted to 1 M KCl and filtered through Superose 6. Fractions (0.4 ml) were collected from 6 to 22 ml. The positions of elution of blue dextran 2000, thyroglobulin (T; 669 kDa), ferritin (F; 443 kDa), catalase (C; 232 kDa), and aldolase (A; 158 kDa) were determined by elution of protein standards. (B) The indicated fractions across the peak of UV absorbance were separated by electrophoresis on an SDS–8% polyacrylamide gel and visualized by staining with Coomassie blue. Lane 2, the load; lanes 3 to 9, fractions 25 to 37, odd numbers only. The position of P143 is indicated on the left. The migration of protein molecular weight markers is indicated on the left. (C) Aliquots of the corresponding fractions were dialyzed to 50 mM KCl, and 2 μl of each was assayed for ATPase activity. Background activity from a no-enzyme control was subtracted.
FIG. 5
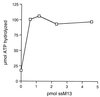
Effect of DNA on P143 ATPase activity. ATPase assay contained 140 fmol of P143 and the indicated amounts of ssM13mp18 per reaction. Each point represents the average of three experiments.
FIG. 6
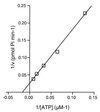
Km of P143 ATP hydrolysis. The rate of ATP hydrolysis was determined for 7.8125, 15.625, 31.25, 62.5, and 125 mM ATP concentrations in the presence of 1 pmol of ssM13 DNA. Reaction mixtures were incubated with 140 fmol of purified P143 for 5 min at 37°C. A double-reciprocal plot of the rate of 32Pi formation versus the ATP concentration is shown.
FIG. 7
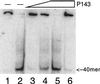
DNA helicase activity of P143. Purified P143 was tested for helicase activity by using the oligonucleotide displacement assay. The substrate consists of an ssM13 molecule with a radiolabeled 40-nucleotide primer annealed; there was 6.75 fmol of substrate per lane. Lane 1, substrate in the absence of enzyme; lane 2, substrate after boiling for 3 min; lanes 3, 4, and 5, 70, 140, and 280 fmol of P143, respectively. The reaction mixture shown in lane 6 was incubated with 280 fmol of P143 in the absence of ATP. Samples were fractionated on a 12% acrylamide–TBE gel. The positions of the free 40-mer are indicated on the right.
FIG. 8
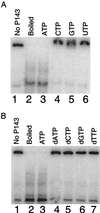
Nucleotide requirement for DNA unwinding activity of P143. Reaction conditions were the same as described in the legend for Fig. 7. (A) Nucleotide specificity of the helicase activity of P143. Lane 1, substrate alone; lane 2, boiled substrate alone; lanes 3 to 6, 280 fmol of P143 with a 10 mM concentration of the indicated recombinant NTP. (B) Sugar specificity of helicase activity. Lanes 3 to 7 contain 280 fmol of P143 with a 10 mM concentration of the indicated dNTP; the reaction mixture shown in lane 3 was incubated in the presence of 10 mM ATP as a positive control.
Similar articles
- Autographa californica nuclear polyhedrosis virus p143 gene product is a DNA-binding protein.
Laufs S, Lu A, Arrell K, Carstens EB. Laufs S, et al. Virology. 1997 Feb 3;228(1):98-106. doi: 10.1006/viro.1996.8361. Virology. 1997. PMID: 9024814 - A baculovirus single-stranded DNA binding protein, LEF-3, mediates the nuclear localization of the putative helicase P143.
Wu Y, Carstens EB. Wu Y, et al. Virology. 1998 Jul 20;247(1):32-40. doi: 10.1006/viro.1998.9235. Virology. 1998. PMID: 9683569 - Extension of Autographa californica nuclear polyhedrosis virus host range by interspecific replacement of a short DNA sequence in the p143 helicase gene.
Croizier G, Croizier L, Argaud O, Poudevigne D. Croizier G, et al. Proc Natl Acad Sci U S A. 1994 Jan 4;91(1):48-52. doi: 10.1073/pnas.91.1.48. Proc Natl Acad Sci U S A. 1994. PMID: 8278405 Free PMC article.
Cited by
- Host AAA+ ATPase TER94 Plays Critical Roles in Building the Baculovirus Viral Replication Factory and Virion Morphogenesis.
Li Y, Hu L, Chen T, Chang M, Deng F, Hu Z, Wang H, Wang M. Li Y, et al. J Virol. 2020 Feb 28;94(6):e01674-19. doi: 10.1128/JVI.01674-19. Print 2020 Feb 28. J Virol. 2020. PMID: 31896597 Free PMC article. - Genomic sequencing of Troides aeacus nucleopolyhedrovirus (TraeNPV) from golden birdwing larvae (Troides aeacus formosanus) to reveal defective Autographa californica NPV genomic features.
Huang YF, Chen TH, Chang ZT, Wang TC, Lee SJ, Kim JC, Kim JS, Chiu KP, Nai YS. Huang YF, et al. BMC Genomics. 2019 May 27;20(1):419. doi: 10.1186/s12864-019-5713-2. BMC Genomics. 2019. PMID: 31133070 Free PMC article. - Autographa californica Nucleopolyhedrovirus AC141 (Exon0), a Potential E3 Ubiquitin Ligase, Interacts with Viral Ubiquitin and AC66 To Facilitate Nucleocapsid Egress.
Biswas S, Willis LG, Fang M, Nie Y, Theilmann DA. Biswas S, et al. J Virol. 2018 Jan 17;92(3):e01713-17. doi: 10.1128/JVI.01713-17. Print 2018 Feb 1. J Virol. 2018. PMID: 29142135 Free PMC article. - Functional Regulation of an Autographa californica Nucleopolyhedrovirus-Encoded MicroRNA, AcMNPV-miR-1, in Baculovirus Replication.
Zhu M, Wang J, Deng R, Wang X. Zhu M, et al. J Virol. 2016 Jun 24;90(14):6526-6537. doi: 10.1128/JVI.00165-16. Print 2016 Jul 15. J Virol. 2016. PMID: 27147751 Free PMC article. - Genomic sequencing and analysis of Sucra jujuba nucleopolyhedrovirus.
Liu X, Yin F, Zhu Z, Hou D, Wang J, Zhang L, Wang M, Wang H, Hu Z, Deng F. Liu X, et al. PLoS One. 2014 Oct 17;9(10):e110023. doi: 10.1371/journal.pone.0110023. eCollection 2014. PLoS One. 2014. PMID: 25329074 Free PMC article.
References
- Ahrens C H, Rohrmann G F. The DNA polymerase and helicase genes of a baculovirus of Orgyia pseudosugata. J Gen Virol. 1996;77:825–837. - PubMed
- Clark R, Lane D P, Tjian R. Use of monoclonal antibodies as probes of simian virus 40 T antigen ATPase activity. J Biol Chem. 1981;256:11854–11858. - PubMed
- Deng W P, Nickoloff J A. Site-directed mutagenesis of virtually any plasmid by eliminating a unique site. Anal Biochem. 1992;200:81–84. - PubMed
- Evans J T, Rosenblatt G S, Leisy D J, Rohrmann G F. Characterization of the interaction between the baculovirus ssDNA-binding protein (LEF-3) and putative helicase (P143) J Gen Virol. 1999;80:493–500. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources