Presenilin 1 is linked with gamma-secretase activity in the detergent solubilized state - PubMed (original) (raw)
Presenilin 1 is linked with gamma-secretase activity in the detergent solubilized state
Y M Li et al. Proc Natl Acad Sci U S A. 2000.
Abstract
gamma-Secretase is a membrane-associated protease that cleaves within the transmembrane region of amyloid precursor protein to generate the C termini of the two Abeta peptide isoforms, Abeta40 and Abeta42. Here we report the detergent solubilization and partial characterization of gamma-secretase. The activity of solubilized gamma-secretase was measured with a recombinant substrate, C100Flag, consisting largely of the C-terminal fragment of amyloid precursor protein downstream of the beta-secretase cleavage site. Cleavage of C100Flag by gamma-secretase was detected by electrochemiluminescence using antibodies that specifically recognize the Abeta40 or Abeta42 termini. Incubation of C100Flag with HeLa cell membranes or detergent-solubilized HeLa cell membranes generates both the Abeta40 and Abeta42 termini. Recovery of catalytically competent, soluble gamma-secretase critically depends on the choice of detergent; CHAPSO (3-[(3-cholamidopropyl)dimethylammonio]-2-hydroxy-1-propanesulfonate) but not Triton X-100 is suitable. Solubilized gamma-secretase activity is inhibited by pepstatin and more potently by a novel aspartyl protease transition-state analog inhibitor that blocks formation of Abeta40 and Abeta42 in mammalian cells. Upon gel exclusion chromatography, solubilized gamma-secretase activity coelutes with presenilin 1 (PS1) at an apparent relative molecular weight of approximately 2.0 x 10(6). Anti-PS1 antibody immunoprecipitates gamma-secretase activity from the solubilized gamma-secretase preparation. These data suggest that gamma-secretase activity is catalyzed by a PS1-containing macromolecular complex.
Figures
Figure 1
In vitro γ-secretase assay. (A) Schematic representation of the fusion protein substrate, C100Flag, consisting sequentially of an N-terminal Met (M), APP597–695 (the Aβ domain is shown), and the Flag tag (Flag) sequence, and its processing by γ-secretase. The Aβ40- and Aβ42-related products (M-Aβ40 and M-Aβ42, respectively) are detected by ECL using biotinylated 4G8 antibody and the ruthenylated G2–10 or FCA3542 antibodies, respectively. (B) Solubilization of γ-secretase activity by extraction of cellular membranes with CHAPSO detergent. HeLa cell membranes were treated with the indicated CHAPSO concentration, and the samples were centrifuged. The supernatant solutions (filled bars) and resuspended pellets (open bars) were diluted in assay buffer to yield a uniform CHAPSO concentration (0.25%), incubated with C100Flag, and assayed for M-Aβ40 production by ECL. The γ-secretase activities are expressed as % of the total activity (supernatant solution plus pellet) at each detergent concentration. The data are the mean values of two independent experiments. (C) Generation of the Aβ40- and Aβ42-related products from C100Flag by HeLa cell membranes or solubilized γ-secretase. Intact membranes (500 μg/ml) (open bars) or solubilized γ-secretase (125 μg/ml) (filled bars) was incubated in the presence of C100Flag (1.7 μM) and CHAPSO (0.25%) at 37°C for 90 min. The reactions were quenched with RIPA and boiled. The resulting mixtures were centrifuged and the supernatant solutions were assayed for M-Aβ40 and M-Aβ42 by ECL. Standard curves using Aβ40 and Aβ42 were generated to measure production of the corresponding peptides. The data show the M-Aβ40 and M-Aβ42 levels as a % of total M-Aβ40 and M-Aβ42 (mean ± SD, n = 3). The total amount of the Aβ-related species was 556 fmol and 310 fmol for the intact membranes and solubilized γ-secretase, respectively. (D) Mass spectrometric confirmation of the identity of the Aβ40-related product (M-Aβ40) in the in vitro γ-secretase assay using solubilized γ-secretase and C100Flag. SELDI, surface-enhanced laser desorption/ionization.
Figure 2
Characterization of detergent-solubilized γ-secretase activity toward C100Flag. (A) Dependence of M-Aβ40 formation on the C100Flag concentration. The data show the ECL signal after a 90-min incubation at 37°C. (B) Time dependence of M-Aβ40 formation by solubilized γ-secretase. The ECL signals are depicted. (C) pH dependence of solubilized γ-secretase activity. The ECL signals for the generation of M-Aβ40 are shown. The mean values from two independent experiments are shown.
Figure 3
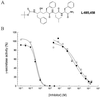
Inhibition of solubilized γ-secretase activity by L-685,458 and pepstatin. (A) Structure of L-685,458. (B) Evaluation of the inhibitory potencies of L-685,458 and pepstatin on solubilized γ-secretase activity. CT100Flag was added to solubilized γ-secretase containing the indicated concentrations of L-685,458 or pepstatin and incubated for 60 min. The ECL assay was used to detect M-Aβ40 (●) and M-Aβ42 (○) in the L-685,458-treated samples and M-Aβ40 (■) and M-Aβ42 (□) in the pepstatin-treated samples. The data are expressed as % of that observed in the absence of either L-685,458 or pepstatin. Shown are the mean values of two and five independent experiments with L-685,458 and pepstatin, respectively.
Figure 4
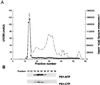
Comigration of γ-secretase activity and PS1 during gel exclusion chromatography of solubilized γ-secretase. (A) Solubilized γ-secretase (1 ml) was chromatographed with Superose 6 gel filtration media. The A280 profile of the eluent is shown (dashed line). Fractions (0.5 ml) were collected and monitored for γ-secretase activity (M-Aβ40 production) with the in vitro γ-secretase assay using C100Flag (●). (B) Column fractions were evaluated for presence of PS1 by SDS/PAGE/immunoblot analysis using anti-PS1-NTF and anti-PS1-CTF antibodies.
Figure 5
Binding of PS1 by immobilized anti-PS1 antibody results in capture of γ-secretase activity. Solubilized γ-secretase was subject to immunoprecipitation (IP) using normal IgG or anti-PS1-NTF IgG. (A) Western blotting analysis using anti-PS1-NTF and anti-PS1-CTF antibodies was performed with the immunoprecipitates. The bands corresponding to PS1-NTF and PS1-CTF are labeled. (B) The resuspended pellets from the normal IgG and anti-PS1 immunoprecipitation steps were evaluated with the in vitro γ-secretase assay (assaying for M-Aβ40 and M-Aβ42). The results are reported as ECL signals (mean of two independent assay determinations).
Comment in
- In search of gamma-secretase: presenilin at the cutting edge.
Selkoe DJ, Wolfe MS. Selkoe DJ, et al. Proc Natl Acad Sci U S A. 2000 May 23;97(11):5690-2. doi: 10.1073/pnas.97.11.5690. Proc Natl Acad Sci U S A. 2000. PMID: 10823929 Free PMC article. No abstract available.
Similar articles
- Aspartyl protease inhibitor pepstatin binds to the presenilins of Alzheimer's disease.
Evin G, Sharples RA, Weidemann A, Reinhard FB, Carbone V, Culvenor JG, Holsinger RM, Sernee MF, Beyreuther K, Masters CL. Evin G, et al. Biochemistry. 2001 Jul 27;40(28):8359-68. doi: 10.1021/bi002770t. Biochemistry. 2001. PMID: 11444983 - gamma-Secretase: characterization and implication for Alzheimer disease therapy.
Xu M, Lai MT, Huang Q, DiMuzio-Mower J, Castro JL, Harrison T, Nadin A, Neduvelil JG, Shearman MS, Shafer JA, Gardell SJ, Li YM. Xu M, et al. Neurobiol Aging. 2002 Nov-Dec;23(6):1023-30. doi: 10.1016/s0197-4580(02)00126-4. Neurobiol Aging. 2002. PMID: 12470798 - In search of gamma-secretase: presenilin at the cutting edge.
Selkoe DJ, Wolfe MS. Selkoe DJ, et al. Proc Natl Acad Sci U S A. 2000 May 23;97(11):5690-2. doi: 10.1073/pnas.97.11.5690. Proc Natl Acad Sci U S A. 2000. PMID: 10823929 Free PMC article. No abstract available. - Secretase as a target for Alzheimer's disease.
Wolfe MS. Wolfe MS. Curr Top Med Chem. 2002 Apr;2(4):371-83. doi: 10.2174/1568026024607535. Curr Top Med Chem. 2002. PMID: 11966461 Review. - gamma-Secretase inhibitors as molecular probes of presenilin function.
Wolfe MS. Wolfe MS. J Mol Neurosci. 2001 Oct;17(2):199-204. doi: 10.1385/JMN:17:2:199. J Mol Neurosci. 2001. PMID: 11816793 Review.
Cited by
- GNG5 is a novel regulator of Aβ42 production in Alzheimer's disease.
Li C, Yang Y, Luo S, Qiu W, Wang X, Ge W. Li C, et al. Cell Death Dis. 2024 Nov 11;15(11):815. doi: 10.1038/s41419-024-07218-z. Cell Death Dis. 2024. PMID: 39528445 Free PMC article. - Alzheimer-mutant γ-secretase complexes stall amyloid β-peptide production.
Arafi P, Devkota S, Maesako M, Wolfe MS. Arafi P, et al. bioRxiv [Preprint]. 2024 Aug 31:2024.08.30.610520. doi: 10.1101/2024.08.30.610520. bioRxiv. 2024. PMID: 39257787 Free PMC article. Preprint. - The aldehyde dehydrogenase 2 rs671 variant enhances amyloid β pathology.
Wang X, Wang J, Chen Y, Qian X, Luo S, Wang X, Ma C, Ge W. Wang X, et al. Nat Commun. 2024 Mar 22;15(1):2594. doi: 10.1038/s41467-024-46899-0. Nat Commun. 2024. PMID: 38519490 Free PMC article. - Familial Alzheimer mutations stabilize synaptotoxic γ-secretase-substrate complexes.
Devkota S, Zhou R, Nagarajan V, Maesako M, Do H, Noorani A, Overmeyer C, Bhattarai S, Douglas JT, Saraf A, Miao Y, Ackley BD, Shi Y, Wolfe MS. Devkota S, et al. Cell Rep. 2024 Feb 27;43(2):113761. doi: 10.1016/j.celrep.2024.113761. Epub 2024 Feb 13. Cell Rep. 2024. PMID: 38349793 Free PMC article. - Genome-wide CRISPR/Cas9 screen identifies regulators of BCMA expression on multiple myeloma cells.
Ajore R, Mattsson J, Pertesi M, Ekdahl L, Ali Z, Hansson M, Nilsson B. Ajore R, et al. Blood Cancer J. 2024 Jan 25;14(1):21. doi: 10.1038/s41408-024-00986-z. Blood Cancer J. 2024. PMID: 38272874 Free PMC article. No abstract available.
References
- Selkoe D J. Trends Cell Biol. 1998;8:447–453. - PubMed
- Jarrett J T, Berger E P, Lansbury P T., Jr Biochemistry. 1993;32:4693–4697. - PubMed
- Kuo Y M, Emmerling M R, Vigo-Pelfrey C, Kasunic T C, Kirkpatrick J B, Murdoch G H, Ball M J, Roher A E. J Biol Chem. 1996;271:4077–4081. - PubMed
- De Strooper B, Annaert W, Cupers P, Saftig P, Craessaerts K, Mumm J S, Schroeter E H, Schrijvers V, Wolfe M S, Ray W J, et al. Nature (London) 1999;398:518–522. - PubMed
- Wolfe M S, Xia W, Moore C L, Leatherwood D D, Ostaszewski B, Rahmati T, Donklor I O, Selkoe D J. Biochemistry. 1999;38:4720–4727. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases