Gene ontology: tool for the unification of biology. The Gene Ontology Consortium - PubMed (original) (raw)
C A Ball, J A Blake, D Botstein, H Butler, J M Cherry, A P Davis, K Dolinski, S S Dwight, J T Eppig, M A Harris, D P Hill, L Issel-Tarver, A Kasarskis, S Lewis, J C Matese, J E Richardson, M Ringwald, G M Rubin, G Sherlock
Affiliations
- PMID: 10802651
- PMCID: PMC3037419
- DOI: 10.1038/75556
Gene ontology: tool for the unification of biology. The Gene Ontology Consortium
M Ashburner et al. Nat Genet. 2000 May.
Abstract
Genomic sequencing has made it clear that a large fraction of the genes specifying the core biological functions are shared by all eukaryotes. Knowledge of the biological role of such shared proteins in one organism can often be transferred to other organisms. The goal of the Gene Ontology Consortium is to produce a dynamic, controlled vocabulary that can be applied to all eukaryotes even as knowledge of gene and protein roles in cells is accumulating and changing. To this end, three independent ontologies accessible on the World-Wide Web (http://www.geneontology.org) are being constructed: biological process, molecular function and cellular component.
Figures
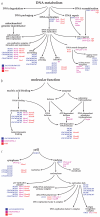
Fig. 1
Examples of Gene Ontology. Three examples illustrate the structure and style used by GO to represent the gene ontologies and to associate genes with nodes within an ontology. The ontologies are built from a structured, controlled vocabulary. The illustrations are the products of work in progress and are subject to change when new evidence becomes available. For simplicity, not all known gene annotations have been included in the figures. a, Biological process ontology. This section illustrates a portion of the biological process ontology describing DNA metabolism. Note that a node may have more than one parent; for example, ‘DNA ligation’ has three parents, ‘DNA-dependent DNA replication’, ‘DNA repair’ and ‘DNA recombination’. b, Molecular function ontology. The ontology is not intended to represent a reaction pathway, but instead reflects conceptual categories of gene-product function. A gene product can be associated with more than one node within an ontology, as illustrated by the MCM proteins. These proteins have been shown to bind chromatin and to possess ATP-dependent DNA helicase activity, and are annotated to both nodes. c, Cellular component ontology. The ontologies are designed for a generic eukaryotic cell, and are flexible enough to represent the known differences between diverse organisms.
Fig. 2
Correspondence between hierarchical clustering of expression microarray experiments with GO terms. The coloured matrix represents the results of clustering many microarray expression experiments. In the matrix, each row represents the yeast gene described to the right, and each column represents the expression of that gene in a particular microarray hybridization. For each gene in the matrix, the table at right lists the systematic ORF name, the standard gene name (if known), and the GO biological process, molecular function and cellular component annotations for that gene. The GO annotations suggest that this experimental expression cluster groups gene products involved in the biological process of protein folding. In contrast, the molecular function and cellular component annotations of these gene products correlate less well with the clustered expression patterns of these gene products.
Similar articles
- A revolution in biology.
Abelson J. Abelson J. Science. 1980 Sep 19;209(4463):1319-21. doi: 10.1126/science.6251541. Science. 1980. PMID: 6251541 No abstract available. - Switching from prokaryotic molecular biology to eukaryotic molecular biology.
Nomura M. Nomura M. J Biol Chem. 2009 Apr 10;284(15):9625-35. doi: 10.1074/jbc.X800014200. Epub 2008 Dec 12. J Biol Chem. 2009. PMID: 19074426 Free PMC article. No abstract available. - Changing tack on the map.
van Heyningen V. van Heyningen V. Nat Genet. 1996 Jun;13(2):134-7. doi: 10.1038/ng0696-134. Nat Genet. 1996. PMID: 8640211 No abstract available. - Molecular biological databases--present and future.
Fuchs R, Rice P, Cameron GN. Fuchs R, et al. Trends Biotechnol. 1992 Jan-Feb;10(1-2):61-6. doi: 10.1016/0167-7799(92)90172-r. Trends Biotechnol. 1992. PMID: 1367938 Review. - Non-coding RNAs: new players in eukaryotic biology.
Costa FF. Costa FF. Gene. 2005 Sep 12;357(2):83-94. doi: 10.1016/j.gene.2005.06.019. Gene. 2005. PMID: 16111837 Review.
Cited by
- Progress and opportunities of foundation models in bioinformatics.
Li Q, Hu Z, Wang Y, Li L, Fan Y, King I, Jia G, Wang S, Song L, Li Y. Li Q, et al. Brief Bioinform. 2024 Sep 23;25(6):bbae548. doi: 10.1093/bib/bbae548. Brief Bioinform. 2024. PMID: 39461902 Free PMC article. Review. - Perturbations in eIF3 subunit stoichiometry alter expression of ribosomal proteins and key components of the MAPK signaling pathways.
Herrmannová A, Jelínek J, Pospíšilová K, Kerényi F, Vomastek T, Watt K, Brábek J, Mohammad MP, Wagner S, Topisirovic I, Valášek LS. Herrmannová A, et al. Elife. 2024 Nov 4;13:RP95846. doi: 10.7554/eLife.95846. Elife. 2024. PMID: 39495207 Free PMC article. - HOXD1 inhibits lung adenocarcinoma progression and is regulated by DNA methylation.
Hu X, Zhang S, Zhang X, Liu H, Diao Y, Li L. Hu X, et al. Oncol Rep. 2024 Dec;52(6):173. doi: 10.3892/or.2024.8832. Epub 2024 Oct 25. Oncol Rep. 2024. PMID: 39450540 Free PMC article. - Acute and chronic impact of interleukin-33 stimulation on chemokines and growth factors in human cord blood-derived mast cells.
Bakhashab S, Banafea GH, Ahmed F, Alsolami R, Schulten HJ, Gauthaman K, Naseer MI, Pushparaj PN. Bakhashab S, et al. PLoS One. 2024 Oct 21;19(10):e0311981. doi: 10.1371/journal.pone.0311981. eCollection 2024. PLoS One. 2024. PMID: 39432538 Free PMC article. - Spatially resolved gene expression profiles of fibrosing interstitial lung diseases.
Kim SJ, Cecchini MJ, Woo E, Jayawardena N, Passos DT, Dick FA, Mura M. Kim SJ, et al. Sci Rep. 2024 Nov 2;14(1):26470. doi: 10.1038/s41598-024-77469-5. Sci Rep. 2024. PMID: 39488596 Free PMC article.
References
- Goffeau A, et al. Life with 6000 genes. Science. 1996;274:546. - PubMed
- Worm Sequencing Consortium. The C. elegans Sequencing Consortium Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998;282:2012–2018. - PubMed
- Adams MD, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. - PubMed
- Meinke DW, et al. Arabidopsis thaliana: a model plant for genome analysis. Science. 1998;282:662–682. - PubMed
Publication types
MeSH terms
Grants and funding
- U41 HG001315/HG/NHGRI NIH HHS/United States
- HD33745/HD/NICHD NIH HHS/United States
- P41 HG001315/HG/NHGRI NIH HHS/United States
- R01 HD033745/HD/NICHD NIH HHS/United States
- P41 HG00330/HG/NHGRI NIH HHS/United States
- R01 HD033745-11/HD/NICHD NIH HHS/United States
- P41 HG000330-22/HG/NHGRI NIH HHS/United States
- U24 HG001315/HG/NHGRI NIH HHS/United States
- P41 HG01315/HG/NHGRI NIH HHS/United States
- P41 HG000739/HG/NHGRI NIH HHS/United States
- P41 HG000330/HG/NHGRI NIH HHS/United States
- U41 HG000739/HG/NHGRI NIH HHS/United States
- P41 HG000739-19/HG/NHGRI NIH HHS/United States
- P41 HG001315-16/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases