Neoplastic transformation by Notch requires nuclear localization - PubMed (original) (raw)
Neoplastic transformation by Notch requires nuclear localization
S Jeffries et al. Mol Cell Biol. 2000 Jun.
Abstract
Notch proteins are plasma membrane-spanning receptors that mediate important cell fate decisions such as differentiation, proliferation, and apoptosis. The mechanism of Notch signaling remains poorly understood. However, it is clear that the Notch signaling pathway mediates its effects through intercellular contact between neighboring cells. The prevailing model for Notch signaling suggests that ligand, presented on a neighboring cell, triggers proteolytic processing of Notch. Following proteolysis, it is thought that the intracellular portion of Notch (N(ic)) translocates to the nucleus, where it is involved in regulating gene expression. There is considerable debate concerning where in the cell Notch functions and what proteins serve as effectors of the Notch signal. Several Notch genes have clearly been shown to be proto-oncogenes in mammalian cells. Activation of Notch proto-oncogenes has been associated with tumorigenesis in several human and other mammalian cancers. Transforming alleles of Notch direct the expression of truncated proteins that primarily consist of N(ic) and are not tethered to the plasma membrane. However, the mechanism by which Notch oncoproteins (generically termed here as N(ic)) induce neoplastic transformation is not known. Previously we demonstrated that N1(ic) and N2(ic) could transform E1A immortalized baby rat kidney cells (RKE) in vitro. We now report direct evidence that N1(ic) must accumulate in the nucleus to induce transformation of RKE cells. In addition, we define the minimal domain of N1(ic) required to induce transformation and present evidence that transformation of RKE cells by N1(ic) is likely to be through a CBF1-independent pathway.
Figures
FIG. 1
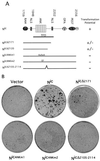
Schematic representation of Nic deletion constructs used in this study. (A) Constructs were designed to delete apparent secondary structure motifs, namely, C-terminal PEST, OPA, and ANK domains and a putative NLS. N-terminal deletions (NicΔR) were designed to remove putative NLSs and the binding site for CBF1 (RAM domain). All constructs have been engineered with a C-terminal Myc epitope tag to facilitate detection (m). The N terminus begins at residue 1759 in Nic constructs and at residue 1848 in NicΔR constructs. Indicated are the residue numbers from full-length human Notch1, which form the C-terminal boundaries of all deletion constructs. Transformation potential refers to the ability of these constructs to elicit focus formation. (B) Verification of Nic deletion mutant expression in transiently transfected Bosc23 cells. Bosc23 cells were transfected with the indicated Nic expression vector as described in Materials and Methods. Cell lysates were analyzed for protein expression by Western blotting against the Myc epitope with 9E10. Indicated above each lane is the residue number that forms the C-terminal boundaries for each deletion construct (top, Nic deletion constructs; bottom, NicΔR deletion constructs). Molecular mass markers are indicated to the left.
FIG. 2
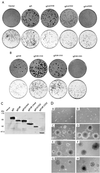
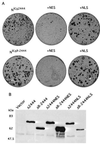
Deletion analysis maps the minimal Nic transforming domain. (A) Nic deletion construct focus assay plates (top row) with corresponding puromycin selection plates (bottom row); (B) NicΔR deletion construct focus assay plates (top row) with corresponding puromycin selection plates (bottom row). Transfected plasmid DNA is indicated above each set of plates (Nic and NicΔR mutant constructs are diagrammed in Fig. 1A). Four weeks posttransfection, the plates were fixed and stained, and foci were counted as described in Materials and Methods. (C) Expression of Nic mutant proteins in RKE cell lines. Transformed RKE cell lysates were immunoprecipitated with Nic polyclonal antiserum 925; proteins were detected by Western blotting with 9E10. (D) Nic-transformed RKE cell lines exhibit anchorage-independent growth in soft agar. Stable RKE cell lines were suspended in 0.35% agarose and monitored for growth; 3 weeks after plating, colonies were photographed at a magnification of ×100. (A) Parental RKE cell line; (B) RKE cell line expressing GFP; (C) Nic, (D) NicΔR; (E) NicΔ-2444; (F) NicΔR-2444; (G) NicΔ2202; (H) NicΔR-2202.
FIG. 3
The TFD is sensitive to mutation. (A) Schematic diagram of additional mutations designed within the TFD (see text). All constructs have been engineered with a C-terminal Myc epitope tag to facilitate detection (m). The N terminus begins at residue 1759 in all Nic mutant constructs; C-terminal boundaries of deletion mutants (NicΔ2171 and NicΔ2120) are indicated. Full-length mutant constructs terminate at reside 2556. Letters (AxAxA in NicANKm1 and EF in NicANKm2) correspond to amino acid substitutions made within the fourth ankyrin repeat; a 10-amino-acid deletion (NicΔ2105–2114) is represented by a gap in full-length Nic. Transformation potential is indicated as + for strong, +/− for weak, and − for negative. (B) Focus assay plates stained 4 weeks after transfection of RKE cells with the indicated Nic mutant constructs.
FIG. 4
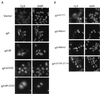
All Nic deletion mutants exhibit nuclear staining regardless of neoplastic properties or NLS deletions. (A) Polyclonal RKE cell lines for each indicated Nic deletion mutant immunostained with 9E10 and visualized with a Cy3-conjugated secondary antibody (left); nuclei are stained with DAPI (right). (B) Nuclear localization is not sufficient for focus formation. Shown are polyclonal RKE cell lines for each indicated Nic deletion or site-directed mutant. NicΔ2171 and site-directed mutant RKE cell lines (NicANKm1, NicANKm2, and NicΔ2105–2114) are immunostained with 9E10 and 15A, respectively, and visualized with a Cy3-conjugated secondary antibody (left); nuclei are indicated by staining with DAPI (right).
FIG. 5
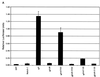
Deletion of the RAM domain from Nic constructs results in loss of CBF1-responsive reporter activity and abolishes binding. (A) HeLa cells were transiently transfected with the indicated Nic expression vector (0.8 μg), 8x-CBF1-luc plasmid (0.4 μg), and RL-TK plasmid (0.4 μg). At 48 h posttransfection, luciferase values were determined for each construct and normalized as described in Materials and Methods (given as relative luciferase units). Shown are the results from one experiment done in triplicate which are representative of multiple assays performed with each deletion construct. (B) NicΔR mutant constructs do not physically associate with GST-CBF1 fusion polypeptide. Cellular lysates were made from stable RKE cell lines expressing the indicated Nic construct. Lysates were incubated with either GST-CBF1 beads (lanes P) or GST beads (lanes C). Nic deletion constructs were detected by Western blotting against the Myc epitope with 9E10. An equivalent amount of each lysate was immunoprecipitated with anti-Notch1 polyclonal antiserum 925 and detected by Western blotting against the Myc epitope (shown in Fig. 2C). Molecular mass markers are shown to the left.
FIG. 5
Deletion of the RAM domain from Nic constructs results in loss of CBF1-responsive reporter activity and abolishes binding. (A) HeLa cells were transiently transfected with the indicated Nic expression vector (0.8 μg), 8x-CBF1-luc plasmid (0.4 μg), and RL-TK plasmid (0.4 μg). At 48 h posttransfection, luciferase values were determined for each construct and normalized as described in Materials and Methods (given as relative luciferase units). Shown are the results from one experiment done in triplicate which are representative of multiple assays performed with each deletion construct. (B) NicΔR mutant constructs do not physically associate with GST-CBF1 fusion polypeptide. Cellular lysates were made from stable RKE cell lines expressing the indicated Nic construct. Lysates were incubated with either GST-CBF1 beads (lanes P) or GST beads (lanes C). Nic deletion constructs were detected by Western blotting against the Myc epitope with 9E10. An equivalent amount of each lysate was immunoprecipitated with anti-Notch1 polyclonal antiserum 925 and detected by Western blotting against the Myc epitope (shown in Fig. 2C). Molecular mass markers are shown to the left.
FIG. 6
Site-directed mutations within the TFD abrogate CBF1 reporter activity without affecting CBF1 binding. (A) HeLa cells were transfected with the indicated Nic mutant construct (see text), and luciferase activity was measured as described in Materials and Methods. (B) Mutations within the TFD do not affect CBF1 binding interactions. Lysates were made from transiently transfected Bosc23 cell lines expressing the indicated Nic constructs and were incubated with either GST-CBF1 beads (lanes P) or GST beads (lanes C). Nic deletion constructs were detected by Western blotting against the Myc epitope with 9E10 (top). Then 10% of each Bosc23 lysate was analyzed for protein expression by Western blotting with 9E10: Nic, lane 1; NicΔR, lane 2; NicANKm1, lane 3; NicANKm2, lane 4 and NicΔ2105–2114, lane 5 (bottom). Molecular mass markers are shown to the left.
FIG. 6
Site-directed mutations within the TFD abrogate CBF1 reporter activity without affecting CBF1 binding. (A) HeLa cells were transfected with the indicated Nic mutant construct (see text), and luciferase activity was measured as described in Materials and Methods. (B) Mutations within the TFD do not affect CBF1 binding interactions. Lysates were made from transiently transfected Bosc23 cell lines expressing the indicated Nic constructs and were incubated with either GST-CBF1 beads (lanes P) or GST beads (lanes C). Nic deletion constructs were detected by Western blotting against the Myc epitope with 9E10 (top). Then 10% of each Bosc23 lysate was analyzed for protein expression by Western blotting with 9E10: Nic, lane 1; NicΔR, lane 2; NicANKm1, lane 3; NicANKm2, lane 4 and NicΔ2105–2114, lane 5 (bottom). Molecular mass markers are shown to the left.
FIG. 7
Nuclear localization is required for Nic-induced transformation of RKE cells. (A) Derivatives of both NicΔ2444 and NicΔR-2444 containing either a PKI-like nuclear export sequence (+NES) or the SV40 large T NLS (+NLS) were constructed and assayed for focus formation in RKE cells as described in Materials and Methods. The transfected expression vector is indicated above each plate. (B) Expression was documented from transiently transfected 293T cells for each indicated construct. Cell lysates were collected and subjected to SDS-PAGE (8% gel), and proteins were visualized by Western blotting with 9E10. Molecular mass markers are shown to the left.
FIG. 8
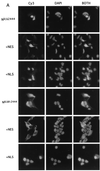
Exclusion from the nucleus correlates with loss of transforming activity. (A) Indicated polyclonal RKE cell lines were processed for IF as described in the text. +NES, clone containing an NES; +NLS, clone containing an NLS. Panels are arranged so that the +NES and +NLS derivatives of the indicated construct follow. Nic constructs are visualized with Cy3 (left); nuclei are stained with DAPI (middle); merged Cy3 and DAPI images are pictured at the right (BOTH). (B) Exclusion from the nucleus depends on active nuclear export. Indicated polyclonal RKE cell lines were plated onto microscope slides and incubated with either the nuclear export inhibitor leptomycin B at 1 ng/ml (+LMB) or with vehicle (−LMB) for 2 h prior to processing for IF. (C) Subcellular localization in transiently transfected HeLa cells. Labels are as indicated for panel A. +NES versions of the construct indicated are to the right.
FIG. 8
Exclusion from the nucleus correlates with loss of transforming activity. (A) Indicated polyclonal RKE cell lines were processed for IF as described in the text. +NES, clone containing an NES; +NLS, clone containing an NLS. Panels are arranged so that the +NES and +NLS derivatives of the indicated construct follow. Nic constructs are visualized with Cy3 (left); nuclei are stained with DAPI (middle); merged Cy3 and DAPI images are pictured at the right (BOTH). (B) Exclusion from the nucleus depends on active nuclear export. Indicated polyclonal RKE cell lines were plated onto microscope slides and incubated with either the nuclear export inhibitor leptomycin B at 1 ng/ml (+LMB) or with vehicle (−LMB) for 2 h prior to processing for IF. (C) Subcellular localization in transiently transfected HeLa cells. Labels are as indicated for panel A. +NES versions of the construct indicated are to the right.
FIG. 8
Exclusion from the nucleus correlates with loss of transforming activity. (A) Indicated polyclonal RKE cell lines were processed for IF as described in the text. +NES, clone containing an NES; +NLS, clone containing an NLS. Panels are arranged so that the +NES and +NLS derivatives of the indicated construct follow. Nic constructs are visualized with Cy3 (left); nuclei are stained with DAPI (middle); merged Cy3 and DAPI images are pictured at the right (BOTH). (B) Exclusion from the nucleus depends on active nuclear export. Indicated polyclonal RKE cell lines were plated onto microscope slides and incubated with either the nuclear export inhibitor leptomycin B at 1 ng/ml (+LMB) or with vehicle (−LMB) for 2 h prior to processing for IF. (C) Subcellular localization in transiently transfected HeLa cells. Labels are as indicated for panel A. +NES versions of the construct indicated are to the right.
Similar articles
- Characterization of a high-molecular-weight Notch complex in the nucleus of Notch(ic)-transformed RKE cells and in a human T-cell leukemia cell line.
Jeffries S, Robbins DJ, Capobianco AJ. Jeffries S, et al. Mol Cell Biol. 2002 Jun;22(11):3927-41. doi: 10.1128/MCB.22.11.3927-3941.2002. Mol Cell Biol. 2002. PMID: 11997524 Free PMC article. - p300 acts as a transcriptional coactivator for mammalian Notch-1.
Oswald F, Täuber B, Dobner T, Bourteele S, Kostezka U, Adler G, Liptay S, Schmid RM. Oswald F, et al. Mol Cell Biol. 2001 Nov;21(22):7761-74. doi: 10.1128/MCB.21.22.7761-7774.2001. Mol Cell Biol. 2001. PMID: 11604511 Free PMC article. - Notch(ic)-ER chimeras display hormone-dependent transformation, nuclear accumulation, phosphorylation and CBF1 activation.
Ronchini C, Capobianco AJ. Ronchini C, et al. Oncogene. 2000 Aug 10;19(34):3914-24. doi: 10.1038/sj.onc.1203719. Oncogene. 2000. PMID: 10951584 - Neoplastic transformation by Notch is independent of transcriptional activation by RBP-J signalling.
Dumont E, Fuchs KP, Bommer G, Christoph B, Kremmer E, Kempkes B. Dumont E, et al. Oncogene. 2000 Jan 27;19(4):556-61. doi: 10.1038/sj.onc.1203352. Oncogene. 2000. PMID: 10698525 - Notch signaling as a target in multimodality cancer therapy.
Jang MS, Zlobin A, Kast WM, Miele L. Jang MS, et al. Curr Opin Mol Ther. 2000 Feb;2(1):55-65. Curr Opin Mol Ther. 2000. PMID: 11249652 Review.
Cited by
- The Esophageal Organoid System Reveals Functional Interplay Between Notch and Cytokines in Reactive Epithelial Changes.
Kasagi Y, Chandramouleeswaran PM, Whelan KA, Tanaka K, Giroux V, Sharma M, Wang J, Benitez AJ, DeMarshall M, Tobias JW, Hamilton KE, Falk GW, Spergel JM, Klein-Szanto AJ, Rustgi AK, Muir AB, Nakagawa H. Kasagi Y, et al. Cell Mol Gastroenterol Hepatol. 2018 Jan 3;5(3):333-352. doi: 10.1016/j.jcmgh.2017.12.013. eCollection 2018 Mar. Cell Mol Gastroenterol Hepatol. 2018. PMID: 29552622 Free PMC article. - Mutations in NOTCH2 in patients with Hajdu-Cheney syndrome.
Zhao W, Petit E, Gafni RI, Collins MT, Robey PG, Seton M, Miller KK, Mannstadt M. Zhao W, et al. Osteoporos Int. 2013 Aug;24(8):2275-81. doi: 10.1007/s00198-013-2298-5. Epub 2013 Feb 7. Osteoporos Int. 2013. PMID: 23389697 Free PMC article. - Notch signaling induces rapid degradation of achaete-scute homolog 1.
Sriuranpong V, Borges MW, Strock CL, Nakakura EK, Watkins DN, Blaumueller CM, Nelkin BD, Ball DW. Sriuranpong V, et al. Mol Cell Biol. 2002 May;22(9):3129-39. doi: 10.1128/MCB.22.9.3129-3139.2002. Mol Cell Biol. 2002. PMID: 11940670 Free PMC article. - Characterization of a high-molecular-weight Notch complex in the nucleus of Notch(ic)-transformed RKE cells and in a human T-cell leukemia cell line.
Jeffries S, Robbins DJ, Capobianco AJ. Jeffries S, et al. Mol Cell Biol. 2002 Jun;22(11):3927-41. doi: 10.1128/MCB.22.11.3927-3941.2002. Mol Cell Biol. 2002. PMID: 11997524 Free PMC article. - Cellular senescence checkpoint function determines differential Notch1-dependent oncogenic and tumor-suppressor activities.
Kagawa S, Natsuizaka M, Whelan KA, Facompre N, Naganuma S, Ohashi S, Kinugasa H, Egloff AM, Basu D, Gimotty PA, Klein-Szanto AJ, Bass AJ, Wong KK, Diehl JA, Rustgi AK, Nakagawa H. Kagawa S, et al. Oncogene. 2015 Apr 30;34(18):2347-59. doi: 10.1038/onc.2014.169. Epub 2014 Jun 16. Oncogene. 2015. PMID: 24931169 Free PMC article.
References
- Ahmad I, Zagouras P, Artavanis-Tsakonas S. Involvement of Notch-1 in mammalian retinal neurogenesis: association of Notch-1 activity with both immature and terminally differentiated cells. Mech Dev. 1995;53:73–85. - PubMed
- Artavanis-Tsakonas S, Rand M D, Lake R J. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. - PubMed
- Aster J, Pear W, Hasserjian R, Erba H, Davi F, Luo B, Scott M, Baltimore D, Sklar J. Functional analysis of the TAN-1 gene, a human homolog of Drosophila notch. Cold Spring Harbor Symp Quant Biol. 1994;59:125–136. - PubMed
- Aster J C, Robertson E S, Hasserjian R P, Turner J R, Kieff E, Sklar J. Oncogenic forms of NOTCH1 lacking either the primary binding site for RBP-Jkappa or nuclear localization sequences retain the ability to associate with RBP-Jkappa and activate transcription. J Biol Chem. 1997;272:11336–11343. - PubMed
- Axelrod J D, Matsuno K, Artavanis-Tsakonas S, Perrimon N. Interaction between Wingless and Notch signaling pathways mediated by dishevelled. Science. 1996;271:1826–1832. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources