Telomere maintenance in telomerase-deficient mouse embryonic stem cells: characterization of an amplified telomeric DNA - PubMed (original) (raw)
Telomere maintenance in telomerase-deficient mouse embryonic stem cells: characterization of an amplified telomeric DNA
H Niida et al. Mol Cell Biol. 2000 Jun.
Abstract
Telomere dynamics, chromosomal instability, and cellular viability were studied in serial passages of mouse embryonic stem (ES) cells in which the telomerase RNA (mTER) gene was deleted. These cells lack detectable telomerase activity, and their growth rate was reduced after more than 300 divisions and almost zero after 450 cell divisions. After this growth crisis, survivor cells with a rapid growth rate did emerge. Such survivors were found to maintain functional telomeres in a telomerase-independent fashion. Although telomerase-independent telomere maintenance has been reported for some immortalized mammalian cells, its molecular mechanism has not been elucidated. Characterization of the telomeric structures in one of the survivor mTER(-/-) cell lines showed amplification of the same tandem arrays of telomeric and nontelomeric sequences at most of the chromosome ends. This evidence implicates cis/trans amplification as one mechanism for the telomerase-independent maintenance of telomeres in mammalian cells.
Figures
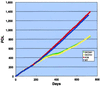
FIG. 1
Growth of _mTER_−/− ES cells in long-term culture. The growth characteristics of mTER+/+ (WT), _mTER_−/+ (KO6), and _mTER_−/− (DKO301 and DKO741) ES cells were monitored during long-term culture.
FIG. 2
Telomerase activity of _mTER_−/− survivor ES cells. Total-cell lysates were prepared from WT, DKO301 at 663 PDL, and DKO741 at 638 PDL. The assay was performed multiple times, and both DKO301 and DKO741 survivors still exhibited undetectable levels of telomerase activity. IC, internal control.
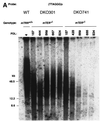
FIG. 3
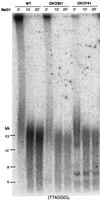
Telomere dynamics in _mTER_−/− ES cells. Genomic DNAs from mTER+/+ (WT) and _mTER_−/− (DKO301 and DKO741) ES cells at selected PDL were digested with _Hin_fI and separated on a pulsed-field gel (A) or a 0.8% agarose gel (B) or digested with _Hap_II and separated on a pulsed-field gel (E). Genomic DNAs from WT cells at 4 PDL, DKO301 cells at 824 PDL, and DKO741 cells at 764 PDL were digested with each 4- or 5-base restriction endonuclease and separated on a pulsed-field gel (C) or a 0.8% agarose gel (D and F). The 5′-[32P](T2AG3)3 telomeric DNA oligonucleotides (A to E) and 555-bp nontelomeric DNA fragment (F) were used as probes.
FIG. 3
Telomere dynamics in _mTER_−/− ES cells. Genomic DNAs from mTER+/+ (WT) and _mTER_−/− (DKO301 and DKO741) ES cells at selected PDL were digested with _Hin_fI and separated on a pulsed-field gel (A) or a 0.8% agarose gel (B) or digested with _Hap_II and separated on a pulsed-field gel (E). Genomic DNAs from WT cells at 4 PDL, DKO301 cells at 824 PDL, and DKO741 cells at 764 PDL were digested with each 4- or 5-base restriction endonuclease and separated on a pulsed-field gel (C) or a 0.8% agarose gel (D and F). The 5′-[32P](T2AG3)3 telomeric DNA oligonucleotides (A to E) and 555-bp nontelomeric DNA fragment (F) were used as probes.
FIG. 3
Telomere dynamics in _mTER_−/− ES cells. Genomic DNAs from mTER+/+ (WT) and _mTER_−/− (DKO301 and DKO741) ES cells at selected PDL were digested with _Hin_fI and separated on a pulsed-field gel (A) or a 0.8% agarose gel (B) or digested with _Hap_II and separated on a pulsed-field gel (E). Genomic DNAs from WT cells at 4 PDL, DKO301 cells at 824 PDL, and DKO741 cells at 764 PDL were digested with each 4- or 5-base restriction endonuclease and separated on a pulsed-field gel (C) or a 0.8% agarose gel (D and F). The 5′-[32P](T2AG3)3 telomeric DNA oligonucleotides (A to E) and 555-bp nontelomeric DNA fragment (F) were used as probes.
FIG. 3
Telomere dynamics in _mTER_−/− ES cells. Genomic DNAs from mTER+/+ (WT) and _mTER_−/− (DKO301 and DKO741) ES cells at selected PDL were digested with _Hin_fI and separated on a pulsed-field gel (A) or a 0.8% agarose gel (B) or digested with _Hap_II and separated on a pulsed-field gel (E). Genomic DNAs from WT cells at 4 PDL, DKO301 cells at 824 PDL, and DKO741 cells at 764 PDL were digested with each 4- or 5-base restriction endonuclease and separated on a pulsed-field gel (C) or a 0.8% agarose gel (D and F). The 5′-[32P](T2AG3)3 telomeric DNA oligonucleotides (A to E) and 555-bp nontelomeric DNA fragment (F) were used as probes.
FIG. 3
Telomere dynamics in _mTER_−/− ES cells. Genomic DNAs from mTER+/+ (WT) and _mTER_−/− (DKO301 and DKO741) ES cells at selected PDL were digested with _Hin_fI and separated on a pulsed-field gel (A) or a 0.8% agarose gel (B) or digested with _Hap_II and separated on a pulsed-field gel (E). Genomic DNAs from WT cells at 4 PDL, DKO301 cells at 824 PDL, and DKO741 cells at 764 PDL were digested with each 4- or 5-base restriction endonuclease and separated on a pulsed-field gel (C) or a 0.8% agarose gel (D and F). The 5′-[32P](T2AG3)3 telomeric DNA oligonucleotides (A to E) and 555-bp nontelomeric DNA fragment (F) were used as probes.
FIG. 3
Telomere dynamics in _mTER_−/− ES cells. Genomic DNAs from mTER+/+ (WT) and _mTER_−/− (DKO301 and DKO741) ES cells at selected PDL were digested with _Hin_fI and separated on a pulsed-field gel (A) or a 0.8% agarose gel (B) or digested with _Hap_II and separated on a pulsed-field gel (E). Genomic DNAs from WT cells at 4 PDL, DKO301 cells at 824 PDL, and DKO741 cells at 764 PDL were digested with each 4- or 5-base restriction endonuclease and separated on a pulsed-field gel (C) or a 0.8% agarose gel (D and F). The 5′-[32P](T2AG3)3 telomeric DNA oligonucleotides (A to E) and 555-bp nontelomeric DNA fragment (F) were used as probes.
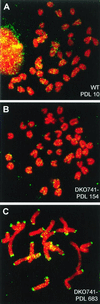
FIG. 4
Metaphase spreads from WT and _mTER_−/− ES cells. FISH with a PNA-telomere probe for WT (A and B), DKO301 (C to E), and DKO741 (F to H) at selected PDL. Two-color FISH with the telomere (green) and mouse major satellite (pink) probes for WT (I) at 1,134 PDL and DKO741 (J) at 683 PDL. Chromosomal stability and telomeric DNA content have been highly sustained in the WT over 1,000 PDL. On the other hand, telomeric DNA content in the _mTER_−/− ES cells has decreased progressively until the growth crisis stage (C and D; F and G) but regained after the crisis by telomerase-independent mechanisms (E and H). Furthermore, many fusions (mostly p-arm-to-p-arm fusions) were induced during the growth crisis stage (D and G).
FIG. 4
Metaphase spreads from WT and _mTER_−/− ES cells. FISH with a PNA-telomere probe for WT (A and B), DKO301 (C to E), and DKO741 (F to H) at selected PDL. Two-color FISH with the telomere (green) and mouse major satellite (pink) probes for WT (I) at 1,134 PDL and DKO741 (J) at 683 PDL. Chromosomal stability and telomeric DNA content have been highly sustained in the WT over 1,000 PDL. On the other hand, telomeric DNA content in the _mTER_−/− ES cells has decreased progressively until the growth crisis stage (C and D; F and G) but regained after the crisis by telomerase-independent mechanisms (E and H). Furthermore, many fusions (mostly p-arm-to-p-arm fusions) were induced during the growth crisis stage (D and G).
FIG. 4
Metaphase spreads from WT and _mTER_−/− ES cells. FISH with a PNA-telomere probe for WT (A and B), DKO301 (C to E), and DKO741 (F to H) at selected PDL. Two-color FISH with the telomere (green) and mouse major satellite (pink) probes for WT (I) at 1,134 PDL and DKO741 (J) at 683 PDL. Chromosomal stability and telomeric DNA content have been highly sustained in the WT over 1,000 PDL. On the other hand, telomeric DNA content in the _mTER_−/− ES cells has decreased progressively until the growth crisis stage (C and D; F and G) but regained after the crisis by telomerase-independent mechanisms (E and H). Furthermore, many fusions (mostly p-arm-to-p-arm fusions) were induced during the growth crisis stage (D and G).
FIG. 4
Metaphase spreads from WT and _mTER_−/− ES cells. FISH with a PNA-telomere probe for WT (A and B), DKO301 (C to E), and DKO741 (F to H) at selected PDL. Two-color FISH with the telomere (green) and mouse major satellite (pink) probes for WT (I) at 1,134 PDL and DKO741 (J) at 683 PDL. Chromosomal stability and telomeric DNA content have been highly sustained in the WT over 1,000 PDL. On the other hand, telomeric DNA content in the _mTER_−/− ES cells has decreased progressively until the growth crisis stage (C and D; F and G) but regained after the crisis by telomerase-independent mechanisms (E and H). Furthermore, many fusions (mostly p-arm-to-p-arm fusions) were induced during the growth crisis stage (D and G).
FIG. 4
Metaphase spreads from WT and _mTER_−/− ES cells. FISH with a PNA-telomere probe for WT (A and B), DKO301 (C to E), and DKO741 (F to H) at selected PDL. Two-color FISH with the telomere (green) and mouse major satellite (pink) probes for WT (I) at 1,134 PDL and DKO741 (J) at 683 PDL. Chromosomal stability and telomeric DNA content have been highly sustained in the WT over 1,000 PDL. On the other hand, telomeric DNA content in the _mTER_−/− ES cells has decreased progressively until the growth crisis stage (C and D; F and G) but regained after the crisis by telomerase-independent mechanisms (E and H). Furthermore, many fusions (mostly p-arm-to-p-arm fusions) were induced during the growth crisis stage (D and G).
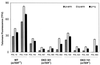
FIG. 5
Q-FISH analysis. Mean telomere fluorescence of mTER+/+ (WT) and _mTER_−/− (DKO301 and DKO741) cells at selected PDL. Fluorescence is expressed in TFU, where 1 TFU corresponds to 1 kb of T2AG3 repeat in plasmid DNA (36). For the survivors (DKO301 at 692 PDL and DKO741 at 683 PDL), the q-arm TFU show a more appropriate telomere length at the ends of chromosomes. Error bars indicate the SE.
FIG. 6
Sequencing and structure of the 1.6-kb _Hin_fI TRF from the DKO741 survivor ES cells. Genomic DNA from DKO741 at PDL 860 was digested with _Hin_fI. Fragments of about 1.6 kb were isolated and subcloned. (A) Nucleotide sequence of the cloned gene. (B) Structure of the cloned 1.6-kb _Hin_fI fragment. Open and shaded boxes indicate telomeric and nontelomeric DNA regions, respectively. The recognition sites of the 4- and 5-base restriction endonucleases used in Fig. 2 are indicated: A, _Alu_I; HIII, _Hae_III; M, _Mbo_I; Hi, _Hin_fI; R, _Rsa_I. The sizes of the restriction fragments are shown. The numbering of restriction sites 3′ to the telomeric region is based on the 3′-most _Hin_fI site as base 1600. With the T2AG3 probe, we expected fragment sizes of 868, 1,315, >1,070, 1,600, and >1,432 bp with _Alu_I, _Hae_III, _Mbo_I, _Hin_fI, and _Rsa_I, respectively. With the 555-bp nontelomeric DNA probe, we expected fragment sizes of 320 and >232, 1,315, >302 and 231, 1,600, and >1,432, respectively, for the same enzymes.
FIG. 7
FISH analysis of nontelomeric DNA sequence. FISH with the 555-bp nontelomeric DNA probe used for Fig. 3F (lower panel). (A) WT cells at 10 PDL; (B) DKO741 at 154 PDL; (C) DKO741 at 683 PDL.
FIG. 8
BAL-31 nuclease treatment assay. Genomic DNA (5 μg) from WT at 1,008 PDL, DKO301 at 663 PDL, or DKO741 at 638 PDL was digested with BAL-31 nuclease for 0, 10, or 20 min. Then, DNA was further digested with _Hap_II and used for TRF analysis. (T2AG3)3 was used as the probe.
Similar articles
- Severe growth defect in mouse cells lacking the telomerase RNA component.
Niida H, Matsumoto T, Satoh H, Shiwa M, Tokutake Y, Furuichi Y, Shinkai Y. Niida H, et al. Nat Genet. 1998 Jun;19(2):203-6. doi: 10.1038/580. Nat Genet. 1998. PMID: 9620783 - The telomerase reverse transcriptase is limiting and necessary for telomerase function in vivo.
Liu Y, Snow BE, Hande MP, Yeung D, Erdmann NJ, Wakeham A, Itie A, Siderovski DP, Lansdorp PM, Robinson MO, Harrington L. Liu Y, et al. Curr Biol. 2000 Nov 16;10(22):1459-62. doi: 10.1016/s0960-9822(00)00805-8. Curr Biol. 2000. PMID: 11102810 - DNA repair factors and telomere-chromosome integrity in mammalian cells.
Hande MP. Hande MP. Cytogenet Genome Res. 2004;104(1-4):116-22. doi: 10.1159/000077475. Cytogenet Genome Res. 2004. PMID: 15162024 Review. - The role of telomeres and telomerase complex in haematological neoplasia: the length of telomeres as a marker of carcinogenesis and prognosis of disease.
Gancarcíková M, Zemanová Z, Brezinová J, Berková A, Vcelíková S, Smigová J, Michalová K. Gancarcíková M, et al. Prague Med Rep. 2010;111(2):91-105. Prague Med Rep. 2010. PMID: 20653999 Review. - Telomeres, telomerase and chromosome stability.
Preston RJ. Preston RJ. Radiat Res. 1997 May;147(5):529-34. Radiat Res. 1997. PMID: 9146697 Review.
Cited by
- A 'higher order' of telomere regulation: telomere heterochromatin and telomeric RNAs.
Schoeftner S, Blasco MA. Schoeftner S, et al. EMBO J. 2009 Aug 19;28(16):2323-36. doi: 10.1038/emboj.2009.197. Epub 2009 Jul 23. EMBO J. 2009. PMID: 19629032 Free PMC article. Review. - Zscan4 regulates telomere elongation and genomic stability in ES cells.
Zalzman M, Falco G, Sharova LV, Nishiyama A, Thomas M, Lee SL, Stagg CA, Hoang HG, Yang HT, Indig FE, Wersto RP, Ko MS. Zalzman M, et al. Nature. 2010 Apr 8;464(7290):858-63. doi: 10.1038/nature08882. Epub 2010 Mar 24. Nature. 2010. PMID: 20336070 Free PMC article. - Loss of function of Atrx leads to activation of alternative lengthening of telomeres in a primary mouse model of sarcoma.
Pierpoint M, Floyd W, Wisdom AJ, Luo L, Ma Y, Waitkus MS, Kirsch DG. Pierpoint M, et al. bioRxiv [Preprint]. 2023 Nov 6:2023.11.06.565874. doi: 10.1101/2023.11.06.565874. bioRxiv. 2023. PMID: 37986934 Free PMC article. Updated. Preprint. - The Saccharomyces cerevisiae WRN homolog Sgs1p participates in telomere maintenance in cells lacking telomerase.
Johnson FB, Marciniak RA, McVey M, Stewart SA, Hahn WC, Guarente L. Johnson FB, et al. EMBO J. 2001 Feb 15;20(4):905-13. doi: 10.1093/emboj/20.4.905. EMBO J. 2001. PMID: 11179234 Free PMC article. - Telomere Biology-Insights into an Intriguing Phenomenon.
Venkatesan S, Khaw AK, Hande MP. Venkatesan S, et al. Cells. 2017 Jun 19;6(2):15. doi: 10.3390/cells6020015. Cells. 2017. PMID: 28629193 Free PMC article. Review.
References
- Bilaud T, Brun C, Ancelin K, Koering C E, Laroche T, Gilson E. Telomeric localization of TRF2, a novel human telobox protein. Nat Genet. 1997;17:236–239. - PubMed
- Blackburn E H. Structure and function of telomeres. Nature. 1991;350:569–573. - PubMed
- Blasco M A, Funk W D, Villeponteau B, Greider C W. Functional characterization and developmental regulation of mouse telomerase RNA. Science. 1995;269:1267–1270. - PubMed
- Blasco M A, Lee H W, Hande M P, Samper E, Lansdorp P M, DePinho R A, Greider C W. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell. 1997;91:25–34. - PubMed
- Bodnar A G, Ouellette M, Frolkis M, Holt S E, Chiu C P, Morin G B, Harley C B, Shay J W, Lichtsteiner S, Wright W E. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279:349–352. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources