Asparagine-proline sequence within membrane-spanning segment of SREBP triggers intramembrane cleavage by site-2 protease - PubMed (original) (raw)
Asparagine-proline sequence within membrane-spanning segment of SREBP triggers intramembrane cleavage by site-2 protease
J Ye et al. Proc Natl Acad Sci U S A. 2000.
Abstract
The NH(2)-terminal domains of membrane-bound sterol regulatory element-binding proteins (SREBPs) are released into the cytosol by regulated intramembrane proteolysis, after which they enter the nucleus to activate genes encoding lipid biosynthetic enzymes. Intramembrane proteolysis is catalyzed by Site-2 protease (S2P), a hydrophobic zinc metalloprotease that cleaves SREBPs at a membrane-embedded leucine-cysteine bond. In the current study, we use domain-swapping methods to localize the residues within the SREBP-2 membrane-spanning segment that are required for cleavage by S2P. The studies reveal a requirement for an asparagine-proline sequence in the middle third of the transmembrane segment. We propose a model in which the asparagine-proline sequence serves as an NH(2)-terminal cap for a portion of the transmembrane alpha-helix of SREBP, allowing the remainder of the alpha-helix to unwind partially to expose the peptide bond for cleavage by S2P.
Figures
Figure 1
Replacement of the first transmembrane domain of SREBP-2 with that of GPP130 abolishes Site-2 cleavage. The DNA sequence encoding the first transmembrane domain of SREBP-2 (designated wild-type) was replaced with DNA encoding the membrane-spanning segment of GPP130 (designated chimera A). The bar denotes the first transmembrane domain with relevant amino acids numbered above. Black and red letters represent SREBP-2 and GPP130 sequences, respectively. M19 cells were set up on day 0 and were transfected on day 1 with a plasmid encoding wild-type or chimeric HSV-tagged SREBP-2 (3 μg/dish) together with pCMV-SCAP(D443N) (0.5 μg/dish) with or without pCMV-Myc-S2P (1 μg/dish) as indicated. The total amount of DNA was adjusted to 4.5 μg/dish by addition of pcDNA3 empty vector. After incubation at 37°C with sterols for 20 h, the cells were harvested and fractionated as described in Materials and Methods. Aliquots of nuclear extracts or membranes (30 μg protein) were subjected to SDS/PAGE and immunoblot analysis as described in Materials and Methods. Filters were exposed for 15 sec. P, I, and N denote the precursor, intermediate, and nuclear forms of HSV-tagged SREBP-2, respectively.
Figure 2
Site-2 cleavage of SREBP-2/GPP130 chimeric proteins in transfected M19 cells. The sequence flanking the first transmembrane domain of wild-type SREBP-2 and various GPP130 chimeric substitutions is shown. The bar denotes the putative first transmembrane domain. Black and red letters represent amino acids derived from SREBP-2 and GPP130, respectively. M19 cells were set up, were transfected with the indicated plasmids, were incubated for 20 h at 37°C with sterols, and were harvested for immunoblot analysis as described in the legend to Fig. 1. Filters were exposed to film for 30 sec in A and 1 min in B. P, I, and N denote the precursor, intermediate, and nuclear forms of HSV-tagged SREBP-2, respectively. The asterisk (*) denotes a cross-reactive band that is present in mock-transfected cells.
Figure 3
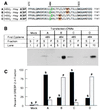
NP sequence in the first transmembrane domain of SREBP-2 is required for cleavage at Site-2. (A) M19 cells were set up, were transfected with the indicated plasmids, were incubated for 20 h at 37°C with sterols, and were harvested for immunoblot analysis as described in the legend to Fig. 1. Filters were exposed to film for 30 sec. P, I, and N denote the precursor, intermediate, and nuclear forms of HSV-tagged SREBP-2, respectively. The asterisk (*) denotes a cross-reactive band that is present in mock-transfected cells. (B) Sequence alignment of the first transmembrane domain of SREBPs from various species. Completely conserved amino acids are boxed. The sequences of human SREBP-1 (28), human SREBP-2 (29), hamster SREBP-1 (30), hamster SREBP-2 (31), and Drosophila SREBP (32) are published in the indicated references. The C. elegans sequence is predicted from the sequence of a cosmid that was obtained from the European Molecular Biology Laboratory Database (ID code AL031635.1).
Figure 4
Movement of NP sequence fails to alter site of cleavage by Site-2 protease as measured by cysteine panning. (A) Sequences of His8-tagged ACBP/SREBP-2 chimeric proteins in which positions of cysteine (green) and asparagine-proline (NP) (yellow) sequences have been moved. The bar above the sequences denotes the first transmembrane domain. Proline-470 is the first amino acid derived from SREBP-2. The first cysteine in the chimeric proteins is placed at a position corresponding to residue 484 or 485 of the SREBP-2 sequence. The NP sequence is highlighted in yellow. (B) Immunoblots of the cleaved proteins after derivatization by _N_-biotinylaminoethyl methanethiosulfonate and precipitation by streptavidin-agarose. Monolayers of HEK-293 cells were transfected with 4 μg of the indicated plasmid together with 0.5 μg of pCMV-SCAP(D443N), 0.5 μg pVAI, and 0.25 μg of pCMV-Myc-S2P. After incubation at 37°C for 20 h without sterols, the cells were harvested, cytosol was prepared, and the cleaved NH2-terminal domains in the cytosolic fraction were purified by Ni-NTA chromatography, were treated with _N_-biotinylaminoethyl methanethiosulfonate, and were precipitated with streptavidin-agarose as described in Materials and Methods. Equal aliquots (50% of total volume of the streptavidin-agarose supernatant (S) and pellet (P) fractions) were subjected to SDS/PAGE and were immunoblotted with IgG-HSV-Tag. The filter was exposed to film for 20 sec at room temperature. (C) Quantification of the bands in A as determined by densitometry as described in Materials and Methods. Each bar represents the percentage of total ACBP/SREBP-2 found in the supernatant (closed bars) or pellet (hatched bars). The results shown are representative of two experiments.
Figure 5
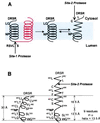
Proposed model for the effect of NP on the conformation of the first transmembrane domain of SREBP-2. (A) The first (black) and second (red) transmembrane domains of SREBP-2 are shown. The NP motif, S1P cleavage site (RSVL↑S), S2P cleavage site (L↓C), and the DRSR sequence immediately adjacent to the first transmembrane domain are shown. After cleavage at Site-1, the first transmembrane domain separates from the second transmembrane domain. This allows the NH2-terminal portion of the transmembrane α-helix to unwind to form a more extended structure, thereby exposing the Leu-Cys (LC) bond to the cytosolic face. The unwinding of the helix requires the NP sequence, which caps the unwinding, allowing the COOH-terminal portion of the transmembrane domain to maintain a helical conformation. (B) Proposed dimensions of the first transmembrane domain in relation to the membrane thickness. The transmembrane domain at the left is presented as a kinked α-helix. The spacing between every turn of the helix is assumed to be 1.5 Å (33), and the thickness of the hydrocarbon region of the bilayer is assumed to be 30 Å (34). On the right, the NH2-terminal end of the transmembrane domain is shown in an extended conformation followed by an α-helix that is capped by the NP motif.
Similar articles
- Membrane topology of S2P, a protein required for intramembranous cleavage of sterol regulatory element-binding proteins.
Zelenski NG, Rawson RB, Brown MS, Goldstein JL. Zelenski NG, et al. J Biol Chem. 1999 Jul 30;274(31):21973-80. doi: 10.1074/jbc.274.31.21973. J Biol Chem. 1999. PMID: 10419520 - Complementation cloning of S2P, a gene encoding a putative metalloprotease required for intramembrane cleavage of SREBPs.
Rawson RB, Zelenski NG, Nijhawan D, Ye J, Sakai J, Hasan MT, Chang TY, Brown MS, Goldstein JL. Rawson RB, et al. Mol Cell. 1997 Dec;1(1):47-57. doi: 10.1016/s1097-2765(00)80006-4. Mol Cell. 1997. PMID: 9659902 - Regulated cleavage of sterol regulatory element binding proteins requires sequences on both sides of the endoplasmic reticulum membrane.
Hua X, Sakai J, Brown MS, Goldstein JL. Hua X, et al. J Biol Chem. 1996 Apr 26;271(17):10379-84. doi: 10.1074/jbc.271.17.10379. J Biol Chem. 1996. PMID: 8626610 - A proteolytic pathway that controls the cholesterol content of membranes, cells, and blood.
Brown MS, Goldstein JL. Brown MS, et al. Proc Natl Acad Sci U S A. 1999 Sep 28;96(20):11041-8. doi: 10.1073/pnas.96.20.11041. Proc Natl Acad Sci U S A. 1999. PMID: 10500120 Free PMC article. Review. - Regulated intramembrane proteolysis: from the endoplasmic reticulum to the nucleus.
Rawson RB. Rawson RB. Essays Biochem. 2002;38:155-68. doi: 10.1042/bse0380155. Essays Biochem. 2002. PMID: 12463168 Review.
Cited by
- OCIAD1 is a host mitochondrial substrate of the hepatitis C virus NS3-4A protease.
Tran HTL, Morikawa K, Anggakusuma, Zibi R, Thi VLD, Penin F, Heim MH, Quadroni M, Pietschmann T, Gouttenoire J, Moradpour D. Tran HTL, et al. PLoS One. 2020 Jul 22;15(7):e0236447. doi: 10.1371/journal.pone.0236447. eCollection 2020. PLoS One. 2020. PMID: 32697788 Free PMC article. - The intramembrane cleavage site of the amyloid precursor protein depends on the length of its transmembrane domain.
Lichtenthaler SF, Beher D, Grimm HS, Wang R, Shearman MS, Masters CL, Beyreuther K. Lichtenthaler SF, et al. Proc Natl Acad Sci U S A. 2002 Feb 5;99(3):1365-70. doi: 10.1073/pnas.032395699. Epub 2002 Jan 22. Proc Natl Acad Sci U S A. 2002. PMID: 11805291 Free PMC article. - Substrate requirements for regulated intramembrane proteolysis of Bacillus subtilis pro-sigmaK.
Prince H, Zhou R, Kroos L. Prince H, et al. J Bacteriol. 2005 Feb;187(3):961-71. doi: 10.1128/JB.187.3.961-971.2005. J Bacteriol. 2005. PMID: 15659674 Free PMC article. - Alternative processing of sterol regulatory element binding protein during larval development in Drosophila melanogaster.
Matthews KA, Kunte AS, Tambe-Ebot E, Rawson RB. Matthews KA, et al. Genetics. 2009 Jan;181(1):119-28. doi: 10.1534/genetics.108.093450. Epub 2008 Nov 17. Genetics. 2009. PMID: 19015545 Free PMC article. - The role of L1 loop in the mechanism of rhomboid intramembrane protease GlpG.
Wang Y, Maegawa S, Akiyama Y, Ha Y. Wang Y, et al. J Mol Biol. 2007 Dec 7;374(4):1104-13. doi: 10.1016/j.jmb.2007.10.014. Epub 2007 Oct 11. J Mol Biol. 2007. PMID: 17976648 Free PMC article.
References
- Brown M S, Ye J, Rawson R B, Goldstein J L. Cell. 2000;100:391–398. - PubMed
- Hua X, Nohturfft A, Goldstein J L, Brown M S. Cell. 1996;87:415–426. - PubMed
- Sakai J, Rawson R B, Espenshade P J, Cheng D, Seegmiller A C, Goldstein J L, Brown M S. Mol Cell. 1998;2:505–514. - PubMed
- Duncan E A, Davé U P, Sakai J, Goldstein J L, Brown M S. J Biol Chem. 1998;273:17801–17809. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous