Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a - PubMed (original) (raw)
Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a
B H Ramsahoye et al. Proc Natl Acad Sci U S A. 2000.
Abstract
Current evidence indicates that methylation of cytosine in mammalian DNA is restricted to both strands of the symmetrical sequence CpG, although there have been sporadic reports that sequences other than CpG may also be methylated. We have used a dual-labeling nearest neighbor technique and bisulphite genomic sequencing methods to investigate the nearest neighbors of 5-methylcytosine residues in mammalian DNA. We find that embryonic stem cells, but not somatic tissues, have significant cytosine-5 methylation at CpA and, to a lesser extent, at CpT. As the expression of the de novo methyltransferase Dnmt3a correlates well with the presence of non-CpG methylation, we asked whether Dnmt3a might be responsible for this modification. Analysis of genomic methylation in transgenic Drosophila expressing Dnmt3a reveals that Dnmt3a is predominantly a CpG methylase but also is able to induce methylation at CpA and at CpT.
Figures
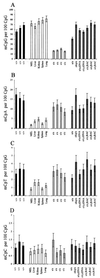
Figure 1
mCpA and mCpT are severalfold more prevalent in wild-type ES cells than in tissues, even in the absence of Dnmt1. Bar charts to illustrate levels of mCpG (A), mCpA (B), mCpT (C), and mCpC levels (D) as measured by NNA. Black bars indicate wild-type ES cells; clear bars indicate Dnmt1-null ES cells; light-gray bars indicate somatic DNAs; and checkered bars indicate Dnmt1 partial mutant, Dnmt1 rescued, and Dnmt1 overexpressing ES cells (see Table 1 for explanation of genotypes and references).
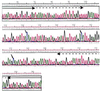
Figure 2
Bisulphite genomic sequencing confirms the presence of non-CpG methylation in ES cell DNA. Sequence of J1–22, a PCR product derived from a bisulphite-modified _Rsa_I fragment from wild-type ES cells (Dnmt1+/+) by using primers directed toward the linker DNA. The small arrows indicate the positions of nonsymmetrically methylated cytosine residues. The large solid arrows indicate the modified _Eco_RI linker DNA sequences. The dashed arrows indicate the sequences to which primers were designed for specific amplification of this DNA segment from bisulphite-modified but unlinked DNA.
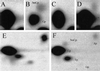
Figure 3
Dnmt3a induces CpG, CpA, and CpT methylation in Drosophila. NNA of genomic DNA extracted from wild-type (A, C, and E) and _Dnmt3a_-transgenic (B, D, and F) Drosophila. (A and B) _Fok_I-digested DNAs labeled with [α-32P]dGTP. (C and D) _Fok_I-digested DNA labeled with [α-32P]dATP. (E and F) _Mva_I-digested DNAs labeled with [α-32P]dATP. In the _Mva_I experiments (E and F), a small amount of labeling Ap, Tp, and Gp can also be seen. This finding is attributable to background labeling at nicks in the DNA. The identities of the labeled nucleotides are shown in B and F.
Figure 4
Model for the reestablishment of DNA methylation after implantation. (A) In early preimplantation development, the genome becomes substantially demethylated. Dnmt1 is transcribed and is active but is not targeted to the nucleus (26). Nothing is known about the activities of Dnmt3a and Dnmt3b in this early stage of development (hence the dotted lines), the earliest stage at which transcripts have been documented being embryonic day 6.5 (20). (B) Northern blot analysis indicates that Dnmt3a activity is particularly high at embryonic day 10.5 (20), and it is reasonable to assume that this finding correlates with Dnmt3a activity. Hence, at this stage, de novo methylation at CpG, CpA, and CpT (square symbols) will be occurring as well as maintenance of mCpG sites by Dnmt1 (round symbols). (C) In late development, Dnmt3 activity is low. Therefore, maintenance methylation predominates, and non-CpG methylation is diminished.
Similar articles
- De novo methylation of MMLV provirus in embryonic stem cells: CpG versus non-CpG methylation.
Dodge JE, Ramsahoye BH, Wo ZG, Okano M, Li E. Dodge JE, et al. Gene. 2002 May 1;289(1-2):41-8. doi: 10.1016/s0378-1119(02)00469-9. Gene. 2002. PMID: 12036582 - CpG sites preferentially methylated by Dnmt3a in vivo.
Oka M, Rodić N, Graddy J, Chang LJ, Terada N. Oka M, et al. J Biol Chem. 2006 Apr 14;281(15):9901-8. doi: 10.1074/jbc.M511100200. Epub 2006 Jan 26. J Biol Chem. 2006. PMID: 16439359 - Control of methylation spreading in synthetic DNA sequences by the murine DNA methyltransferase.
Tollefsbol TO, Hutchison CA 3rd. Tollefsbol TO, et al. J Mol Biol. 1997 Jun 20;269(4):494-504. doi: 10.1006/jmbi.1997.1064. J Mol Biol. 1997. PMID: 9217255 - DNA Methyltransferases in Mammalian Oocytes.
Uysal F, Ozturk S. Uysal F, et al. Results Probl Cell Differ. 2017;63:211-222. doi: 10.1007/978-3-319-60855-6_10. Results Probl Cell Differ. 2017. PMID: 28779320 Review. - Mediating and maintaining methylation while minimizing mutation: Recent advances on mammalian DNA methyltransferases.
Cheng X, Blumenthal RM. Cheng X, et al. Curr Opin Struct Biol. 2022 Aug;75:102433. doi: 10.1016/j.sbi.2022.102433. Epub 2022 Jul 29. Curr Opin Struct Biol. 2022. PMID: 35914495 Free PMC article. Review.
Cited by
- Epigenetic regulation of cardiovascular diseases induced by behavioral and environmental risk factors: Mechanistic, diagnostic, and therapeutic insights.
Bi F, Gao C, Guo H. Bi F, et al. FASEB Bioadv. 2024 Oct 1;6(11):477-502. doi: 10.1096/fba.2024-00080. eCollection 2024 Nov. FASEB Bioadv. 2024. PMID: 39512842 Free PMC article. Review. - Using human disease mutations to understand de novo DNA methyltransferase function.
Rolls W, Wilson MD, Sproul D. Rolls W, et al. Biochem Soc Trans. 2024 Oct 30;52(5):2059-2075. doi: 10.1042/BST20231017. Biochem Soc Trans. 2024. PMID: 39446312 Free PMC article. Review. - DNA methylation analysis to differentiate reference, breed, and parent-of-origin effects in the bovine pangenome era.
MacPhillamy C, Chen T, Hiendleder S, Williams JL, Alinejad-Rokny H, Low WY. MacPhillamy C, et al. Gigascience. 2024 Jan 2;13:giae061. doi: 10.1093/gigascience/giae061. Gigascience. 2024. PMID: 39435573 Free PMC article. - Environmental exposures influence multigenerational epigenetic transmission.
Klibaner-Schiff E, Simonin EM, Akdis CA, Cheong A, Johnson MM, Karagas MR, Kirsh S, Kline O, Mazumdar M, Oken E, Sampath V, Vogler N, Wang X, Nadeau KC. Klibaner-Schiff E, et al. Clin Epigenetics. 2024 Oct 17;16(1):145. doi: 10.1186/s13148-024-01762-3. Clin Epigenetics. 2024. PMID: 39420431 Free PMC article. Review. - Complex Pathophysiology of Acute Kidney Injury (AKI) in Aging: Epigenetic Regulation, Matrix Remodeling, and the Healing Effects of H2S.
Gupta S, Mandal S, Banerjee K, Almarshood H, Pushpakumar SB, Sen U. Gupta S, et al. Biomolecules. 2024 Sep 17;14(9):1165. doi: 10.3390/biom14091165. Biomolecules. 2024. PMID: 39334931 Free PMC article. Review.
References
- Sinsheimer R L. J Biol Chem. 1955;215:579–583. - PubMed
- Gruenbaum Y, Stein R, Cedar H, Razin A. FEBS Lett. 1981;124:67–71. - PubMed
- Salomon R, Kaye A. Biochim Biophys Acta. 1970;204:340–351. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials