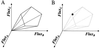The Escherichia coli MG1655 in silico metabolic genotype: its definition, characteristics, and capabilities - PubMed (original) (raw)
The Escherichia coli MG1655 in silico metabolic genotype: its definition, characteristics, and capabilities
J S Edwards et al. Proc Natl Acad Sci U S A. 2000.
Abstract
The Escherichia coli MG1655 genome has been completely sequenced. The annotated sequence, biochemical information, and other information were used to reconstruct the E. coli metabolic map. The stoichiometric coefficients for each metabolic enzyme in the E. coli metabolic map were assembled to construct a genome-specific stoichiometric matrix. The E. coli stoichiometric matrix was used to define the system's characteristics and the capabilities of E. coli metabolism. The effects of gene deletions in the central metabolic pathways on the ability of the in silico metabolic network to support growth were assessed, and the in silico predictions were compared with experimental observations. It was shown that based on stoichiometric and capacity constraints the in silico analysis was able to qualitatively predict the growth potential of mutant strains in 86% of the cases examined. Herein, it is demonstrated that the synthesis of in silico metabolic genotypes based on genomic, biochemical, and strain-specific information is possible, and that systems analysis methods are available to analyze and interpret the metabolic phenotype.
Figures
Figure 1
The feasible solution set for a hypothetical metabolic reaction network. (A) The steady-state operation of the metabolic network is restricted to the region within a cone, defined as the feasible set (8). The feasible set contains all flux vectors that satisfy the physicochemical constrains (Eqs. 1 and 2). Thus, the feasible set defines the capabilities of the metabolic network. All feasible metabolic flux distributions lie within the feasible set, and (B) in the limiting case, where all constraints on the metabolic network are known, such as the enzyme kinetics and gene regulation, the feasible set may be reduced to a single point. This single point must lie within the feasible set.
Figure 2
Gene deletions in E. coli MG1655 central intermediary metabolism; maximal biomass yields on glucose for all possible single gene deletions in the central metabolic pathways. The optimal value of the mutant objective function (Z_mutant) compared with the “wild-type” objective function (Z), where Z is defined in Eq. 3. The ratio of optimal growth yields (Z_mutant/Z). The results were generated in a simulated aerobic environment with glucose as the carbon source. The transport fluxes were constrained as follows: _β_glucose = 10 mmol/g-dry weight (DW) per h; _β_oxygen = 15 mmol/g-DW per h. The maximal yields were calculated by using FBA with the objective of maximizing growth. The biomass yields are normalized with respect to the results for the full metabolic genotype. The yellow bars represent gene deletions that reduced the maximal biomass yield to less than 95% of the in silico wild type.
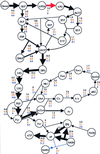
Figure 3
Rerouting of metabolic fluxes. (Black) Flux distribution for the complete gene set. (Red) _zwf_- mutant. Biomass yield is 99% of the results for the full metabolic genotype. (Blue) _zwf_- _pnt_- mutant. Biomass yield is 92% of the results for the full metabolic genotype (see text). The solid lines represent enzymes that are being used, with the corresponding flux value noted. The fluxes [substrates converted/h per g-dry weight (DW)] were calculated by using FBA with the input parameters of glucose uptake rate (_β_glucose = 6.6 mmol glucose/h per g-DW) and oxygen uptake rate (_β_oxygen = 12.4 mmol oxygen/h per g-DW) (41).
Similar articles
- In silico predictions of Escherichia coli metabolic capabilities are consistent with experimental data.
Edwards JS, Ibarra RU, Palsson BO. Edwards JS, et al. Nat Biotechnol. 2001 Feb;19(2):125-30. doi: 10.1038/84379. Nat Biotechnol. 2001. PMID: 11175725 - Metabolic flux balance analysis and the in silico analysis of Escherichia coli K-12 gene deletions.
Edwards JS, Palsson BO. Edwards JS, et al. BMC Bioinformatics. 2000;1:1. doi: 10.1186/1471-2105-1-1. Epub 2000 Jul 27. BMC Bioinformatics. 2000. PMID: 11001586 Free PMC article. - Toward metabolic phenomics: analysis of genomic data using flux balances.
Schilling CH, Edwards JS, Palsson BO. Schilling CH, et al. Biotechnol Prog. 1999 May-Jun;15(3):288-95. doi: 10.1021/bp9900357. Biotechnol Prog. 1999. PMID: 10356245 - Constraining the metabolic genotype-phenotype relationship using a phylogeny of in silico methods.
Lewis NE, Nagarajan H, Palsson BO. Lewis NE, et al. Nat Rev Microbiol. 2012 Feb 27;10(4):291-305. doi: 10.1038/nrmicro2737. Nat Rev Microbiol. 2012. PMID: 22367118 Free PMC article. Review. - Escherichia coli proteome analysis using the gene-protein database.
VanBogelen RA, Abshire KZ, Moldover B, Olson ER, Neidhardt FC. VanBogelen RA, et al. Electrophoresis. 1997 Aug;18(8):1243-51. doi: 10.1002/elps.1150180805. Electrophoresis. 1997. PMID: 9298644 Review.
Cited by
- Systems biology beyond degree, hubs and scale-free networks: the case for multiple metrics in complex networks.
Roy S. Roy S. Syst Synth Biol. 2012 Jun;6(1-2):31-4. doi: 10.1007/s11693-012-9094-y. Epub 2012 May 29. Syst Synth Biol. 2012. PMID: 23730362 Free PMC article. - MicrobesFlux: a web platform for drafting metabolic models from the KEGG database.
Feng X, Xu Y, Chen Y, Tang YJ. Feng X, et al. BMC Syst Biol. 2012 Aug 2;6:94. doi: 10.1186/1752-0509-6-94. BMC Syst Biol. 2012. PMID: 22857267 Free PMC article. - Inferring metabolic states in uncharacterized environments using gene-expression measurements.
Rossell S, Huynen MA, Notebaart RA. Rossell S, et al. PLoS Comput Biol. 2013;9(3):e1002988. doi: 10.1371/journal.pcbi.1002988. Epub 2013 Mar 21. PLoS Comput Biol. 2013. PMID: 23555222 Free PMC article. - Advances in metabolic flux analysis toward genome-scale profiling of higher organisms.
Basler G, Fernie AR, Nikoloski Z. Basler G, et al. Biosci Rep. 2018 Nov 23;38(6):BSR20170224. doi: 10.1042/BSR20170224. Print 2018 Dec 21. Biosci Rep. 2018. PMID: 30341247 Free PMC article. Review. - From bit to it: how a complex metabolic network transforms information into living matter.
Wagner A. Wagner A. BMC Syst Biol. 2007 Jul 30;1:33. doi: 10.1186/1752-0509-1-33. BMC Syst Biol. 2007. PMID: 17663759 Free PMC article.
References
- Bailey J E. Biotechnol Prog. 1998;14:8–20. - PubMed
- Kacser H, Burns J A. Symp Soc Exp Biol. 1973;27:65–104. - PubMed
- Fell D. Understanding the Control of Metabolism. London: Portland; 1996.
- Varma A, Palsson B O. Bio/Technology. 1994;12:994–998.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources