Differences in genotypes of Helicobacter pylori from different human populations - PubMed (original) (raw)
Comparative Study
. 2000 Jun;182(11):3210-8.
doi: 10.1128/JB.182.11.3210-3218.2000.
A K Mukhopadhyay, B Velapatiño, W Su, Z Pan, C Garcia, V Hernandez, Y Valdez, R S Mistry, R H Gilman, Y Yuan, H Gao, T Alarcón, M López-Brea, G Balakrish Nair, A Chowdhury, S Datta, M Shirai, T Nakazawa, R Ally, I Segal, B C Wong, S K Lam, F O Olfat, T Borén, L Engstrand, O Torres, R Schneider, J E Thomas, S Czinn, D E Berg
Affiliations
- PMID: 10809702
- PMCID: PMC94509
- DOI: 10.1128/JB.182.11.3210-3218.2000
Comparative Study
Differences in genotypes of Helicobacter pylori from different human populations
D Kersulyte et al. J Bacteriol. 2000 Jun.
Abstract
DNA motifs at several informative loci in more than 500 strains of Helicobacter pylori from five continents were studied by PCR and sequencing to gain insights into the evolution of this gastric pathogen. Five types of deletion, insertion, and substitution motifs were found at the right end of the H. pylori cag pathogenicity island. Of the three most common motifs, type I predominated in Spaniards, native Peruvians, and Guatemalan Ladinos (mixed Amerindian-European ancestry) and also in native Africans and U.S. residents; type II predominated among Japanese and Chinese; and type III predominated in Indians from Calcutta. Sequences in the cagA gene and in vacAm1 type alleles of the vacuolating cytotoxin gene (vacA) of strains from native Peruvians were also more like those from Spaniards than those from Asians. These indications of relatedness of Latin American and Spanish strains, despite the closer genetic relatedness of Amerindian and Asian people themselves, lead us to suggest that H. pylori may have been brought to the New World by European conquerors and colonists about 500 years ago. This thinking, in turn, suggests that H. pylori infection might have become widespread in people quite recently in human evolution.
Figures
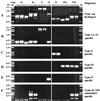
FIG. 1
PCR amplification to detect and identify different insertion, deletion, and substitution motifs at the extreme right end of the cag PAI. The primers used in these amplifications are listed in Table 4, and their positions are diagrammed in Fig. 2. The same DNAs were used for each of the six PCR tests shown here, and in most cases their sequences in this region are known. The strains used (Genbank accession numbers or references) are as follows (from left to right): OhioM6 (AF190992), Gambia94/24 (AF190658), Peru002B (AF190994) (all type Ia); India120A (AF191014) and NTCT11638 (20) (type Ib); 84-183 (AF190660) and OhioP46 (AF190995 and AF190996) (type Ic); 26695 (56) (type IV); JapanHU38 (AF191000); Peruvian Hp1 (from an ethnic Japanese in Peru) (AF190661) (type II); NCTC11637 (AF191010); India17A (AF191016) (type IIIa); and India75A (AF190663) and India68A (not sequenced) (type IIIb). (A) Amplification of entire segment between cagA and glr using primers 1 and 3 (Table 4; Fig. 2). The difference in PCR product size between the two type IIIa strains is due to a 29-bp deletion in NCTC11637, relative to India17A. (B) Type I- and IV-specific amplification using primers 3 and 4 (Table 4; Fig. 2). (C) Type II-specific amplification using primers 3 and 5 (Table 4; Fig. 2). (D) Type III-specific amplification using primers 3 and 6 (Table 4; Fig. 2). (E) Type IV-specific amplification using primers 1 and 7 (Table 4; Fig. 2). (F) MiniIS_605_-specific amplification using primers 1 and 8 (Table 4; Fig. 2) (type Ib or IIIb).
FIG. 2
Diagram of DNA sequence motif types at the right end of the cag PAI of H. pylori and positions of primers that were most useful for distinguishing different motif types and subtypes. RJ, right junction, the 31 bp that contains the 3′ end of the glutamate racemase gene (glr) and that is also repeated directly at the left end of the cag PAI (2, 20, 56). The cagA gene is relatively near the right end of the cag PAI. Closely related sequences in different motif types are indicated by common symbols. The sequences that led to the interpretations diagrammed here have been deposited under GenBank accession no. AF190658, AF190992 to AF190994, and AF191013 (type Ia); AF190659 and AF191014 (type Ib); AF190660, AF190995, and AF190996 (type Ic); AF190661 and AF190997 to AF1901008 (type II); AF190662, AF191009 to AF191012, AF191015, and AF191016 (type IIIa); AF190663 (type IIIb); and AF200689, AF201074, and AF201075 (type V). The type IV sequence was determined in reference as part of the strain 26695 genome sequencing effort. In terms of other published sequences, NCTC11638 (2, 20) and J99 (6) are of types Ib and Ia, respectively. The ancestral IS_606_* element is inferred to have been about 2 kb long, based on its homologs (e.g., canonical IS_606_ and IS_605_) (39, 56), and the IS_606_* remnants shown here are closely related to corresponding regions at one end of canonical IS_606_, with sequence matches of 82 to 85%. The ancestral form of the DNA hel gene found in this cag PAI right junction region may have been more than 1 kb long, based on the sizes of its homologs in the database (see, e.g., MJ0104 of M. janaschii) (17). It is inferred to have undergone various deletion or substitution events that removed much of its 5′ end and upstream sequences in different lineages. The two remnant forms of this gene were designated omega (in NCTC11638, a type Ia strain) (20) and HP0548 (in 26695, a type IV strain) (56). The ∼200-bp unk1 (function unknown) sequence that replaces nearly all of hel in type III strains is not obviously related to other sequences found to date in GenBank and seems not to be a fragment of any complete open reading frame. When present, the ∼268-bp miniIS_605_ element diagrammed here was always found inserted at the same site in the IS_606_* remnant, just downstream of TTTAA (Fig. 1F and 5). Five of the nine type III strains analyzed by sequencing contained a 39-bp deletion in the 66-bp region between the remnants of IS_606_* and the helicase (hel) gene, starting 3 bp after the IS_606_* deletion breakpoint: ChinaF30A (accession no. AF191009), JapanHU54 (accession no. AF191011), India17A (accession no. AF191016), and NCTC11637 (AF191010; also has an additional 29-bp deletion). Type III strains without this 39-bp deletion include India75A (accession no. 190663), India7A (accession no. AF191015), Sweden53 (accession no. 190662), and Peru466 (accession no. AF191012). One type III strain (India47A; accession no. AF200690) also contained a 158-bp deletion within the unk1 sequence. The three type V strains, all from India, were very similar in sequence.
FIG. 3
DNA sequences at 3′ end of helicase (hel) gene. GenBank accession numbers or references for sequences presented here are as follows: type II strains (A) JapanC7, AF190999; ChinaR30, AF190997; Lith5-1, AF190998; Hp1, AF190661; type I and IV strains (B) NCTC11638, ; 26695, . The data in panel A suggest that just the last three codons at the 3′ end of the hel gene (boldface) are retained in type II strains. Also shown is the 22-bp deletion that was found in all type II strains but not in any type I or IV strains (see also Fig. 2). Corresponding sequences from representative type I and IV strains, which have a much longer hel gene fragment, are shown in panel B.
FIG. 4
DNA sequences that identify the location and termini of the unk1 (function unknown) sequence. GenBank accession numbers or references for these sequences are as follows: AF19009 (ChinaF30A), AF191011 (JapanHU54), AF191015 (India74), AF191012 (Peru466), AF191014 (India20A), AF190994 (Peru2B). The unk1 sequence is in lowercase. It seems to be inserted just after the first codon of the remnant of the hel gene found in type I strains (boldface) and to replace hel gene and downstream sequences almost to the 31-bp direct repeat that marks the right end of the cag PAI. Corresponding sequences from two representative type I strains (which have a much longer hel gene fragment; boldface) are also shown.
FIG. 5
Representative sequences at sites of insertion of miniIS_605_ elements. Representative sequences at termini of miniIS_605_ or full-length canonical IS_605_ are in capitals and boldface. Flanking sequences are in lowercase. (A) Examples of miniIS_605_ insertions at a site in the remnant of IS_606_* in the right junction region of the cag PAI. (B) Corresponding empty sites in strains that lack miniIS_605_ at this site. (C) The ends of canonical IS_605_ and its TTTAA target site sequence (39). GenBank accession numbers or references from which present sequences were extracted are as follows: India75A (a type IIIb strain), AF190663; India120A (a type Ib strain), AF191014; JapanHU54 (a type III strain), AF191011; NCTC11638 (a type Ib strain), ; Gambia9424 (a type Ia strain), AF190658; Lith5-1 (a type II strain), AF190998. PCR tests also indicated that miniIS_605_ was inserted at the same site in all strains of type Ib and IIIb. Note that the leftmost 8 bp of miniIS_605_ are variable in sequence and were initially (20) not recognized as being part of this element.
FIG. 6
PCR analysis of middle-region alleles of vacA (vacuolating cytotoxin gene). Three allelic types were recognized (vacAm1a, -m1b, and -m2) (8, 47). Sizes (0.6 and 0.2 kb) of marker DNAs in the 100-bp ladder are shown. J and S, representative Japanese and Spanish strains, respectively. Other strains for which PCR profiles are shown are from native Peruvians.
Similar articles
- Distinctiveness of genotypes of Helicobacter pylori in Calcutta, India.
Mukhopadhyay AK, Kersulyte D, Jeong JY, Datta S, Ito Y, Chowdhury A, Chowdhury S, Santra A, Bhattacharya SK, Azuma T, Nair GB, Berg DE. Mukhopadhyay AK, et al. J Bacteriol. 2000 Jun;182(11):3219-27. doi: 10.1128/JB.182.11.3219-3227.2000. J Bacteriol. 2000. PMID: 10809703 Free PMC article. - Genomes of Helicobacter pylori from native Peruvians suggest admixture of ancestral and modern lineages and reveal a western type cag-pathogenicity island.
Devi SM, Ahmed I, Khan AA, Rahman SA, Alvi A, Sechi LA, Ahmed N. Devi SM, et al. BMC Genomics. 2006 Jul 27;7:191. doi: 10.1186/1471-2164-7-191. BMC Genomics. 2006. PMID: 16872520 Free PMC article. - Genetic characterisation of Helicobacter pylori isolates from an Argentinean adult population based on cag pathogenicity island right-end motifs, lspA-glmM polymorphism and iceA and vacA genotypes.
Leanza AG, Matteo MJ, Crespo O, Antelo P, Olmos J, Catalano M. Leanza AG, et al. Clin Microbiol Infect. 2004 Sep;10(9):811-9. doi: 10.1111/j.1198-743X.2004.00940.x. Clin Microbiol Infect. 2004. PMID: 15355412 - Helicobacter pylori typing as a tool for tracking human migration.
Yamaoka Y. Yamaoka Y. Clin Microbiol Infect. 2009 Sep;15(9):829-34. doi: 10.1111/j.1469-0691.2009.02967.x. Clin Microbiol Infect. 2009. PMID: 19702588 Free PMC article. Review.
Cited by
- Role of Helicobacter pylori plasticity region genes in development of gastroduodenal diseases.
Sugimoto M, Watada M, Jung SW, Graham DY, Yamaoka Y. Sugimoto M, et al. J Clin Microbiol. 2012 Feb;50(2):441-8. doi: 10.1128/JCM.00906-11. Epub 2011 Nov 23. J Clin Microbiol. 2012. PMID: 22116145 Free PMC article. - Sequential inactivation of rdxA (HP0954) and frxA (HP0642) nitroreductase genes causes moderate and high-level metronidazole resistance in Helicobacter pylori.
Jeong JY, Mukhopadhyay AK, Dailidiene D, Wang Y, Velapatiño B, Gilman RH, Parkinson AJ, Nair GB, Wong BC, Lam SK, Mistry R, Segal I, Yuan Y, Gao H, Alarcon T, Brea ML, Ito Y, Kersulyte D, Lee HK, Gong Y, Goodwin A, Hoffman PS, Berg DE. Jeong JY, et al. J Bacteriol. 2000 Sep;182(18):5082-90. doi: 10.1128/JB.182.18.5082-5090.2000. J Bacteriol. 2000. PMID: 10960091 Free PMC article. - Molecular evolution of the Helicobacter pylori vacuolating toxin gene vacA.
Gangwer KA, Shaffer CL, Suerbaum S, Lacy DB, Cover TL, Bordenstein SR. Gangwer KA, et al. J Bacteriol. 2010 Dec;192(23):6126-35. doi: 10.1128/JB.01081-10. Epub 2010 Sep 24. J Bacteriol. 2010. PMID: 20870762 Free PMC article. - Ethnic difference in serology of Helicobacter pylori CagA between Japanese and non-Japanese Brazilians for non-cardia gastric cancer.
Tatemichi M, Hamada GS, Nishimoto IN, Kowalski LP, Iriya K, Rodrigues JJ, Tsugane S. Tatemichi M, et al. Cancer Sci. 2003 Jan;94(1):64-9. doi: 10.1111/j.1349-7006.2003.tb01353.x. Cancer Sci. 2003. PMID: 12708476 Free PMC article. - Genetic and transmission analysis of Helicobacter pylori strains within a family.
Raymond J, Thiberg JM, Chevalier C, Kalach N, Bergeret M, Labigne A, Dauga C. Raymond J, et al. Emerg Infect Dis. 2004 Oct;10(10):1816-21. doi: 10.3201/eid1010.040042. Emerg Infect Dis. 2004. PMID: 15504269 Free PMC article.
References
- Achtman M, Azuma T, Berg D E, Ito Y, Morelli G, Pan Z J, Suerbaum S, Thompson S A, van der Ende A, van Doorn L J. Recombination and clonal groupings within Helicobacter pylori from different geographical regions. Mol Microbiol. 1999;32:459–470. - PubMed
- Akopyants N S, Clifton S W, Kersulyte D, Crabtree J E, Youree B E, Reece C A, Bukanov N O, Drazek E S, Roe B A, Berg D E. Analyses of the cag pathogenicity island of Helicobacter pylori. Mol Microbiol. 1998;28:37–54. - PubMed
- Allison M J, Bergman T, Gerszten E. Further studies on fecal parasites in antiquity. Am J Clin Pathol. 1999;112:605–609. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical