A complex of mammalian ufd1 and npl4 links the AAA-ATPase, p97, to ubiquitin and nuclear transport pathways - PubMed (original) (raw)
A complex of mammalian ufd1 and npl4 links the AAA-ATPase, p97, to ubiquitin and nuclear transport pathways
H H Meyer et al. EMBO J. 2000.
Abstract
The AAA-ATPase, p97/Cdc48p, has been implicated in many different pathways ranging from membrane fusion to ubiquitin-dependent protein degradation. Binding of the p47 complex directs p97 to act in the post-mitotic fusion of Golgi membranes. We now describe another binding complex comprising mammalian Ufd1 and Npl4. Yeast Ufd1p is required for ubiquitin-dependent protein degradation whereas yeast Npl4p has been implicated in nuclear transport. In rat liver cytosol, Ufd1 and Npl4 form a binary complex, which exists either alone or bound to p97. Ufd1/Npl4 competes with p47 for binding to p97 and so inhibits Golgi membrane fusion. This suggests that it is involved in another cellular function catalysed by p97, the most likely being ubiquitin-dependent events during mitosis. The fact that the binding of p47 and Ufd1/Npl4 is mutually exclusive suggests that these protein complexes act as adapters, directing a basic p97 activity into different cellular pathways.
Figures
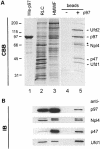
Fig. 1. Affinity purification of cytosolic binding proteins using His-tagged p97. (A) Coomassie Blue-stained gel (CBB) of the purification steps. Recombinant His-tagged p97 (1 µg in lane 1) was biotinylated and immobilized on streptavidin beads. Rat liver cytosol (RLC, 10 µg in lane 2) was fractionated by gel filtration. The high molecular weight fraction (HMWF, 10 µg in lane 3) containing the p97 peak was salt-treated to separate proteins bound to endogenous p97 and then incubated with immobilized His-p97 or streptavidin beads alone. After lowering the salt concentration to permit binding to immobilized p97, the bound proteins were eluted with salt, TCA-precipitated and fractionated using 10% SDS–PAGE (lanes 4 and 5). The bands indicated were excised and analysed by mass spectrometry. Those identified are indicated: p97, p47, Ufd1 (42 kDa), Ufd2 (130 kDa) and a 67 kDa protein with high homology to yeast Npl4p. See text for details. (B) Identification of the p97 binding proteins by immunoblotting (IB). Ten micrograms of RLC, 5 µg HMWF and 5% of the eluates were subjected to SDS–PAGE as decribed in (A). Proteins were transferred to nitrocellulose and incubated with antibodies against p97, p47 and with antibodies generated against Ufd1 and Npl4 (see Figure 3). The anti-Npl4 antibody recognized a doublet of 67 and 71 kDa [asterisk in (A)].
Fig. 2. The rat Npl4 sequence. (A) Nucleotide and predicted amino acid sequence of rat Npl4. Peptide 1 (boxed and shaded) was identified by mass spectrometric analysis of the 67 kDa band and matched two mouse EST clones with high homology to yeast Npl4. The EST sequence was used to screen a rat liver λgt11 cDNA library to isolate the full length rat Npl4 clone shown here. Peptides matching in sequence (shaded) or mass (underlined) with data obtained from the 67 kDa band are indicated. A zinc finger motif is boxed (see text). (B) Rat Npl4 (rat) shares high homology with yeast (S.cer) Npl4p and C.elegans (C. eleg.) gene F59E12.5 throughout the entire sequence (33 and 37% identities, 38 and 29% similarities, respectively), but only rat and the C.elegans sequence contain a zinc finger (ZF). (C) Alignment of homologous zinc fingers of rat Npl4 and C.elegans F59E12.5 with those of RanBP2 and Nup153.
Fig. 2. The rat Npl4 sequence. (A) Nucleotide and predicted amino acid sequence of rat Npl4. Peptide 1 (boxed and shaded) was identified by mass spectrometric analysis of the 67 kDa band and matched two mouse EST clones with high homology to yeast Npl4. The EST sequence was used to screen a rat liver λgt11 cDNA library to isolate the full length rat Npl4 clone shown here. Peptides matching in sequence (shaded) or mass (underlined) with data obtained from the 67 kDa band are indicated. A zinc finger motif is boxed (see text). (B) Rat Npl4 (rat) shares high homology with yeast (S.cer) Npl4p and C.elegans (C. eleg.) gene F59E12.5 throughout the entire sequence (33 and 37% identities, 38 and 29% similarities, respectively), but only rat and the C.elegans sequence contain a zinc finger (ZF). (C) Alignment of homologous zinc fingers of rat Npl4 and C.elegans F59E12.5 with those of RanBP2 and Nup153.
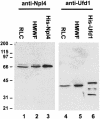
Fig. 3. Expression of recombinant Npl4 and Ufd1 and generation of antibodies. Rat Npl4 and mouse Ufd1 were expressed both as GST fusion proteins and as His-tagged proteins. Antibodies were raised in rabbits against the proteins fused to GST and affinity purified using His-tagged antigens. RLC (10 µg, lanes 1 and 4), HMWF (10 µg, lanes 2 and 5), His-Npl4 (20 ng, lane 3) and His-Ufd1 (20 ng, lane 6) were fractionated using SDS–PAGE and blots probed using anti-Npl4 (lanes 1–3) and anti-Ufd1 (lanes 4–6) antibodies. Anti-Npl4 recognizes a doublet at 67 and 71 kDa in RLC and HMWF. Anti-Ufd1 recognizes a single band at 42 kDa. Both recombinant proteins are slightly bigger than the endogenous proteins because of the His-tag and the His-Ufd1 fraction contains proteolytic degradation products.
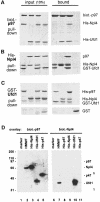
Fig. 4. Complexes of p97, Npl4 and Ufd1. (A–C) Pull down experiments with purified proteins followed by SDS–PAGE and Coomassie Blue staining. The panel on the left shows 10% of input, the right panel the material pulled down. (A) Biotinylated His-p97 was incubated with His-Npl4, His-Ufd1 or His-Npl4/His-Ufd1 and pulled down with streptavidin beads. His-Npl4/His-Ufd1 alone served as the control. (B) His-Npl4 was incubated with GST–Ufd1, rat liver p97 or GST–Ufd1 and rat liver p97 together and pulled down with Ni-NTA beads. GST–Ufd1 and p97 together served as a control. (C) GST–Ufd1 was incubated with His-p97, His-Npl4, or His-p97 and His-Npl4 together and pulled down with glutathione beads. GST alone with His-p97 and His-Npl4 served as the control. Note that more p97 was pulled down in the presence of His-Npl4. (D) Far-Western blots of p97 binding proteins probed with biotinylated p97 or biotinylated Npl4 in the overlay. The cytosolic HMWF (5 µg, lanes 2 and 7) and recombinant proteins as indicated (50 ng in lanes 3–5, 20 ng in lanes 8–11) were fractionated by SDS–PAGE, transferred onto nitrocellulose and overlaid with biotinylated p97 (lanes 1–5) or biotinylated Npl4 (lanes 6–11). Bound probes were visualized using streptavidin-HRP followed by chemiluminescence. p97 recognized recombinant Npl4, Ufd1 and p47, but only Npl4 from cytosol. Npl4 in the overlay bound endogenous Ufd1 in HMWF and the recombinant protein, but not p97, Npl4 or p47. Marker proteins served as a negative control (lanes 1 and 6, myosin, β-galactosidase, phosphorylase b, serum albumin, ovalbumin, carbonic anhydrase, 100 ng each).
Fig. 5. Cytosol contains two independent complexes: a binary Ufd1/Npl4 and a ternary p97/Ufd1/Npl4 complex. (A) Rat liver cytosol (RLC) was fractionated by gel filtration on a Superose 6 column. Aliquots were separated by SDS–PAGE and p97, and its binding proteins analysed by immunoblotting. Almost all the p47 comigrated with the p97 peak at 600–800 kDa. In contrast, Npl4 and Ufd1 ran together exhibiting a biphasic distribution, the first peak co-migrating with p97, the second at ∼200 kDa. (B) Immunoprecipitation of Ufd1 complexes from RLC gel filtration fractions. Total RLC and selected gel filtration fractions identified in (A) were subjected to low stringency immunoprecipitation with a monoclonal anti-Ufd1 antibody. As controls, immunoprecipitation was performed from total RLC using non-immune mouse IgG (lane 1) and from buffer using anti-Ufd1 (lane 7). Precipitates were analysed by SDS–PAGE followed by Coomassie Blue-staining (CBB) or immunoblotting (IB) using specific rabbit antibodies. Npl4 and p97 were both co-immunoprecipitated with Ufd1 from total RLC (lane 2). When precipitated from the high molecular weight peak (lane 4), Ufd1 pulled down both Npl4 and p97, whereas precipitation from the 200 kDa peak (lane 5), bound only Npl4. Note that Ufd1 was not precipitated from fraction 17 (lane 6) where monomeric Ufd1 would be expected. HC, IgG heavy chain; LC, IgG light chain.
Fig. 5. Cytosol contains two independent complexes: a binary Ufd1/Npl4 and a ternary p97/Ufd1/Npl4 complex. (A) Rat liver cytosol (RLC) was fractionated by gel filtration on a Superose 6 column. Aliquots were separated by SDS–PAGE and p97, and its binding proteins analysed by immunoblotting. Almost all the p47 comigrated with the p97 peak at 600–800 kDa. In contrast, Npl4 and Ufd1 ran together exhibiting a biphasic distribution, the first peak co-migrating with p97, the second at ∼200 kDa. (B) Immunoprecipitation of Ufd1 complexes from RLC gel filtration fractions. Total RLC and selected gel filtration fractions identified in (A) were subjected to low stringency immunoprecipitation with a monoclonal anti-Ufd1 antibody. As controls, immunoprecipitation was performed from total RLC using non-immune mouse IgG (lane 1) and from buffer using anti-Ufd1 (lane 7). Precipitates were analysed by SDS–PAGE followed by Coomassie Blue-staining (CBB) or immunoblotting (IB) using specific rabbit antibodies. Npl4 and p97 were both co-immunoprecipitated with Ufd1 from total RLC (lane 2). When precipitated from the high molecular weight peak (lane 4), Ufd1 pulled down both Npl4 and p97, whereas precipitation from the 200 kDa peak (lane 5), bound only Npl4. Note that Ufd1 was not precipitated from fraction 17 (lane 6) where monomeric Ufd1 would be expected. HC, IgG heavy chain; LC, IgG light chain.
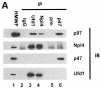
Fig. 6. Ufd1/Npl4 and p47 compete for p97 binding and form alternative complexes in cytosol. (A) Immunoprecipitation of p97 complexes from RLC and analysis of their components. Total RLC was subjected to low stringency immunoprecipitation (IP) using purified IgG, purified antibodies against Ufd1 or Npl4, preimmune-serum (pre-) or anti-serum against p47. Precipitates were fractionated using SDS–PAGE, then analysed by immunoblotting (IB) with antibodies against p97, Npl4, Ufd1 and p47. HMWF (5 µg) was loaded as a reference (lane 1). Note that anti-Ufd1 precipitated Npl4 and p97 but not p47, whereas anti-p47 precipitated p97, but not Npl4 or Ufd1. Rabbit antibodies were used for IP, mouse antibodies for immunodetection, apart from anti-Npl4, which explains the high background. (B) Competition experiments using purified proteins. GST–p47 (lanes 1–12) or GST–Ufd1 with or without Npl4 (lanes 13–20) were incubated with p97 in the absence or presence of increasing amounts of the alternative binding partners (molar excess: 0, 1, 5 and 25 times) as indicated. GST fusion proteins were pulled down with glutathione beads and the bound complexes were analysed by SDS–PAGE followed by Coomassie Blue staining. The lower panel shows 10% of the input, the upper panel shows bound protein. Note the amount of p97 pulled down in each lane.
Fig. 6. Ufd1/Npl4 and p47 compete for p97 binding and form alternative complexes in cytosol. (A) Immunoprecipitation of p97 complexes from RLC and analysis of their components. Total RLC was subjected to low stringency immunoprecipitation (IP) using purified IgG, purified antibodies against Ufd1 or Npl4, preimmune-serum (pre-) or anti-serum against p47. Precipitates were fractionated using SDS–PAGE, then analysed by immunoblotting (IB) with antibodies against p97, Npl4, Ufd1 and p47. HMWF (5 µg) was loaded as a reference (lane 1). Note that anti-Ufd1 precipitated Npl4 and p97 but not p47, whereas anti-p47 precipitated p97, but not Npl4 or Ufd1. Rabbit antibodies were used for IP, mouse antibodies for immunodetection, apart from anti-Npl4, which explains the high background. (B) Competition experiments using purified proteins. GST–p47 (lanes 1–12) or GST–Ufd1 with or without Npl4 (lanes 13–20) were incubated with p97 in the absence or presence of increasing amounts of the alternative binding partners (molar excess: 0, 1, 5 and 25 times) as indicated. GST fusion proteins were pulled down with glutathione beads and the bound complexes were analysed by SDS–PAGE followed by Coomassie Blue staining. The lower panel shows 10% of the input, the upper panel shows bound protein. Note the amount of p97 pulled down in each lane.
Fig. 7. Ufd1/Npl4 does not function as a p97 co-factor in Golgi membrane fusion. (A) MGFs isolated through a 0.5 M sucrose cushion were incubated with p97/p47, p97, p47, p97/Ufd1/Npl4, p97/Ufd1, Npl4 or Ufd1 for 60 min at 37°C. Incubations were processed for EM, and the percentage of membrane in cisternae determined. Results are presented as the percentage cisternal regrowth, where 0% represents starting MGFs and 100% represents the effect of p97/p47. Values represent means ± SEM (n = 3). (B) MGFs were incubated as in (A) with p97/p47 in the presence of increasing amounts of Ufd1 alone or Ufd1/Npl4 (1, 5, 10 and 20 molar excess over p47).
Fig. 7. Ufd1/Npl4 does not function as a p97 co-factor in Golgi membrane fusion. (A) MGFs isolated through a 0.5 M sucrose cushion were incubated with p97/p47, p97, p47, p97/Ufd1/Npl4, p97/Ufd1, Npl4 or Ufd1 for 60 min at 37°C. Incubations were processed for EM, and the percentage of membrane in cisternae determined. Results are presented as the percentage cisternal regrowth, where 0% represents starting MGFs and 100% represents the effect of p97/p47. Values represent means ± SEM (n = 3). (B) MGFs were incubated as in (A) with p97/p47 in the presence of increasing amounts of Ufd1 alone or Ufd1/Npl4 (1, 5, 10 and 20 molar excess over p47).
Fig. 8. Immunofluorescence microscopy showing the localization of Npl4, Ufd1 and p97. Monkey BS-C-1 cells (A, C, E and F) and Normal rat kidney cells (B and D) were stained with affinity-purified antibodies against Npl4 and p97, and a monoclonal antibody against Ufd1 as indicated. Cells were either fixed with paraformaldehyde/methanol (A, C, D and E) or methanol (B and F). C and E is a double staining of Ufd1 and p97 in the same cells. Note that the staining pattern for p97 depends on the fixation method (see text).
Similar articles
- The AAA ATPase p97/VCP interacts with its alternative co-factors, Ufd1-Npl4 and p47, through a common bipartite binding mechanism.
Bruderer RM, Brasseur C, Meyer HH. Bruderer RM, et al. J Biol Chem. 2004 Nov 26;279(48):49609-16. doi: 10.1074/jbc.M408695200. Epub 2004 Sep 15. J Biol Chem. 2004. PMID: 15371428 - Structural insights into the p97-Ufd1-Npl4 complex.
Pye VE, Beuron F, Keetch CA, McKeown C, Robinson CV, Meyer HH, Zhang X, Freemont PS. Pye VE, et al. Proc Natl Acad Sci U S A. 2007 Jan 9;104(2):467-72. doi: 10.1073/pnas.0603408104. Epub 2007 Jan 3. Proc Natl Acad Sci U S A. 2007. PMID: 17202270 Free PMC article. - Direct binding of ubiquitin conjugates by the mammalian p97 adaptor complexes, p47 and Ufd1-Npl4.
Meyer HH, Wang Y, Warren G. Meyer HH, et al. EMBO J. 2002 Nov 1;21(21):5645-52. doi: 10.1093/emboj/cdf579. EMBO J. 2002. PMID: 12411482 Free PMC article. - Cdc48-Ufd1-Npl4: stuck in the middle with Ub.
Bays NW, Hampton RY. Bays NW, et al. Curr Biol. 2002 May 14;12(10):R366-71. doi: 10.1016/s0960-9822(02)00862-x. Curr Biol. 2002. PMID: 12015140 Review. - Expanding into new markets--VCP/p97 in endocytosis and autophagy.
Bug M, Meyer H. Bug M, et al. J Struct Biol. 2012 Aug;179(2):78-82. doi: 10.1016/j.jsb.2012.03.003. Epub 2012 Mar 19. J Struct Biol. 2012. PMID: 22450227 Review.
Cited by
- The role of the N-domain in the ATPase activity of the mammalian AAA ATPase p97/VCP.
Niwa H, Ewens CA, Tsang C, Yeung HO, Zhang X, Freemont PS. Niwa H, et al. J Biol Chem. 2012 Mar 9;287(11):8561-70. doi: 10.1074/jbc.M111.302778. Epub 2012 Jan 23. J Biol Chem. 2012. PMID: 22270372 Free PMC article. - Ubiquitination in the ERAD Process.
Lopata A, Kniss A, Löhr F, Rogov VV, Dötsch V. Lopata A, et al. Int J Mol Sci. 2020 Jul 28;21(15):5369. doi: 10.3390/ijms21155369. Int J Mol Sci. 2020. PMID: 32731622 Free PMC article. Review. - Modulations in the host cell proteome by the hantavirus nucleocapsid protein.
Royster A, Ren S, Ali S, Mir S, Mir M. Royster A, et al. PLoS Pathog. 2024 Jan 8;20(1):e1011925. doi: 10.1371/journal.ppat.1011925. eCollection 2024 Jan. PLoS Pathog. 2024. PMID: 38190410 Free PMC article. - COP9 signalosome interacts ATP-dependently with p97/valosin-containing protein (VCP) and controls the ubiquitination status of proteins bound to p97/VCP.
Cayli S, Klug J, Chapiro J, Fröhlich S, Krasteva G, Orel L, Meinhardt A. Cayli S, et al. J Biol Chem. 2009 Dec 11;284(50):34944-53. doi: 10.1074/jbc.M109.037952. Epub 2009 Oct 13. J Biol Chem. 2009. PMID: 19826004 Free PMC article. - An arginine/lysine-rich motif is crucial for VCP/p97-mediated modulation of ataxin-3 fibrillogenesis.
Boeddrich A, Gaumer S, Haacke A, Tzvetkov N, Albrecht M, Evert BO, Müller EC, Lurz R, Breuer P, Schugardt N, Plassmann S, Xu K, Warrick JM, Suopanki J, Wüllner U, Frank R, Hartl UF, Bonini NM, Wanker EE. Boeddrich A, et al. EMBO J. 2006 Apr 5;25(7):1547-58. doi: 10.1038/sj.emboj.7601043. Epub 2006 Mar 9. EMBO J. 2006. PMID: 16525503 Free PMC article.
References
- Acharya U., Jacobs,R., Peters,J.-U., Watson,N., Farquhar,M.G. and Malhotra,V. (1995) The formation of Golgi stacks from vesiculated Golgi membranes requires two distinct fusion events. Cell, 82, 895–904. - PubMed
- Baldini A. (1999) Is the genetic basis of DiGeorge syndrome in HAND? Nature Genet., 21, 246–247. - PubMed
- Bleasby A.J. and Wootton,J.C. (1990) Construction of validated, non-redundant composite protein sequence databases. Protein Eng., 3, 153–159. - PubMed
- Coles M., Diercks,T., Liermann,J., Groger,A., Rockel,B., Baumeister,W., Koretke,K.K., Lupas,A., Peters,J. and Kessler,H. (1999) The solution structure of VAT-N reveals a ‘missing link’ in the evolution of complex enzymes from a simple βαββ element. Curr. Biol., 9, 1158–1168. - PubMed
- Dai R., Chen,E., Longo,D., Gorbea,C. and Li,C. (1998) Involvement of valosin-containing protein, an ATPase co-purified with IκBα and 26 S proteasome, in ubiquitin-proteasome-mediated degradation of IκBα. J. Biol. Chem., 273, 3562–3573. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous