Transcriptional repression by Pax5 (BSAP) through interaction with corepressors of the Groucho family - PubMed (original) (raw)
Transcriptional repression by Pax5 (BSAP) through interaction with corepressors of the Groucho family
D Eberhard et al. EMBO J. 2000.
Abstract
Pax5 (BSAP) functions as both a transcriptional activator and repressor during midbrain patterning, B-cell development and lymphomagenesis. Here we demonstrate that Pax5 exerts its repression function by recruiting members of the Groucho corepressor family. In a yeast two-hybrid screen, the groucho-related gene product Grg4 was identified as a Pax5 partner protein. Both proteins interact cooperatively via two separate domains: the N-terminal Q and central SP regions of Grg4, and the octapeptide motif and C-terminal transactivation domain of Pax5. The phosphorylation state of Grg4 is altered in vivo upon Pax5 binding. Moreover, Grg4 efficiently represses the transcriptional activity of Pax5 in an octapeptide-dependent manner. Similar protein interactions resulting in transcriptional repression were also observed between distantly related members of both the Pax2/5/8 and Groucho protein families. In agreement with this evolutionary conservation, the octapeptide motif of Pax proteins functions as a Groucho-dependent repression domain in Drosophila embryos. These data indicate that Pax proteins can be converted from transcriptional activators to repressors through interaction with corepressors of the Groucho protein family.
Figures
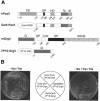
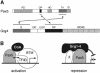
Fig. 1. Interaction of Pax5 with the N-terminal region of Grg4 in yeast. (A) Schematic diagram of Pax5, Grg4 and the respective fusion proteins. The different domains of each protein are indicated together with the corresponding amino acid positions. The chimeric Gal4–Pax5 protein was used as a bait to isolate the VP16-Grg4 fusion protein in a yeast two-hybrid screen of a cDNA expression library that was generated by fusing the transactivation domain (TAD) of VP16 to cDNA derived from 9.5/10.5-day-old mouse embryos (Hollenberg et al., 1995). See text for description of the Grg4 structure. PD, paired domain; OP, octapeptide; HD, partial homeodomain; TA, trans activation region; ID, inhibitory domain; DBD, DNA-binding domain. (B) Specific interaction of Pax5 and Grg4 in yeast. Expression plasmids of the combinations indicated (central panel) were transformed into yeast and selected for by growth on plates lacking leucine and tryptophan (left panel). Activation of a Gal4-dependent HIS3 gene was examined by growth on selection plates additionally lacking histidine (right panel).
Fig. 2. Schematic diagram of mutant Pax5 proteins. (A) Domain structure of Pax5 and extent of amino acid deletion in the different mutant proteins. (B) The octapeptide sequence is shown together with the Y179E mutation and the ΔOP and Δ9A deletions. (C) Conservation of the octapeptide motif. The octapeptide sequences of mouse (m), zebrafish (zf) and Drosophila (d) members of the Pax2/5/8 family are aligned with the corresponding Engrailed homology region 1 (eh1) of the homeodomain transcription factors En2 and Goosecoid (Gsc). Identical amino acids are highlighted by black overlay. For sequences, see Smith and Jaynes (1996), Czerny et al. (1997) and Pfeffer et al. (1998).
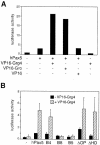
Fig. 3. The N-terminal region of Grg4 interacts with the transactivation domain of Pax5. (A) Interaction of Pax5 and VP16-Grg4. The expression plasmids indicated (100 ng) were co-transfected into J558L cells together with the Renilla luciferase gene pRL-SV40 (0.4 µg) and firefly luciferase gene luc-CD19 (5 µg; Dörfler and Busslinger, 1996). After 48 h, the cells were lysed, luciferase activities were measured, and the activity of the firefly luciferase was standardized relative to the control Renilla luciferase to normalize for differences in transfection efficiencies. Luciferase values are shown relative to the activity measured with the empty expression vector pKW2T (left bar). (B) Interaction of the Q domain of Grg4 with the transactivation domain of Pax5. Both luciferase genes and the Pax5 expression plasmids indicated (100 ng each) were electroporated into J558L cells with (hatched bars) or without (black bars) the VP16-Grg4 expression vector (100 ng). Average values of three experiments are shown.
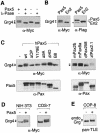
Fig. 4. In vitro binding of Pax5 to Grg4. (A) C-terminal deletions of GST–Grg4 proteins. (B) The SP domain of Grg4 interacts with Pax5. GST pull-down assays were used to study the interaction between in vitro translated, 35S-labeled Pax5 protein and GST (lane 2) or GST–Grg4 proteins (lanes 3–7) bound to glutathione–Sepharose. Lane 1 contained 10% of the Pax5 protein input. (C) The octapeptide of Pax5 mediates binding of Grg4. The 35S-labeled Pax proteins indicated were analyzed for binding to GST or GST–Grg4. The input lane contained ∼10% of the total Pax protein used in each assay.
Fig. 5. In vivo interaction of Groucho and Pax5. Myc-tagged Grg4 or Groucho proteins were expressed in transiently transfected COP-8 fibroblasts either alone (lanes 1, 3 and 5) or with a Flag-tagged Pax5 protein (lanes 2, 4 and 6). After 48 h, whole-cell lysates were prepared and immunoprecipitated with the anti-Flag M2 antibody. Immuno complexes were analyzed by Western blotting first with a polyclonal anti-Myc antibody and then with a polyclonal antiserum directed against the Pax5 paired domain (Adams et al., 1992). Lanes 1–4 contained ∼7.5% of the total input (I) protein or supernatant (S), respectively. P, precipitated protein.
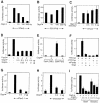
Fig. 6. Induced phosphorylation of Grg4 upon interaction with Pax5. (A) Pax5-dependent phosphorylation of Grg4. Pax5 (where indicated) and Myc-tagged Grg4 were transiently expressed in COP-8 cells for 48 h. Grg4 was then precipitated from whole-cell lysates with a monoclonal anti-Myc antibody, incubated with λ protein phosphatase (λ-Pase) and then analyzed by Western blotting with a polyclonalanti-Myc antibody. (B) Specificity of the Grg4 phosphorylation. Myc-tagged Grg4 and Flag-tagged Pax5 or En2 proteins were co-expressed in COP-8 cells followed by Western blot analysis with anti-Myc and anti-Flag antibodies, respectively. The asterisk denotes a cross-reacting protein. (C) The phosphorylation of Grg4 depends on direct interaction with Pax2/5/8 proteins. The Pax proteins indicated were co-expressed with Myc-tagged Grg4 in COP-8 cells followed by Western blotting with polyclonal anti-Myc and anti-paired domain antibodies. (D) Phosphorylation of Myc-tagged Grg4 in NIH 3T3 and COS-7 cells co-expressing Pax5. (E) Endogenous (endo) Grg proteins are phosphorylated in Pax5-expressing COP-8 cells, as shown by Western blot analysis with a pan-TLE (Grg) antibody (Stifani et al., 1992).
Fig. 7. Grg4 represses the transcriptional activity of Pax5. The Pax expression vector (0.5 µg), luciferase genes luc-CD19 (5 µg) and pRL-SV40 (0.4 µg), and increasing amounts of the indicated Grg expression vector were used for transient transfection of SP2/0 cells (A–D and G–I). The amount of expression plasmid was equalized by the addition of pKW2T, and all data were evaluated as described in the legend to Figure 3A. Normalized luciferase values of one representative experiment (A and B) or three independent transfections (C–I) are shown relative to the luciferase activity measured in the absence of the Pax protein. (A) Repression of the Pax5 transactivation function by Grg4. (B) Failure of Grg4 to repress the function of a paired domain (PD)-VP16 protein. (C) Inability of the Grg4-ΔWD40 protein to repress the transcriptional activity of Pax5. (D) Grg5 fails to antagonize Grg4-mediated repression of Pax5 activity. (E) Repression of a chromatinized reporter gene by Grg4. Expression vectors (1 µg) encoding Gal4–Pax5 or the Gal4 DNA-binding domain (DBD) were transfected together with a Grg4 expression plasmid (0.5 µg) into U2-OS cells containing an integrated luciferase gene, followed by luciferase analysis as described (Alkema et al., 1997). (F) Grg4-mediated repression of the rat TPO promoter. HeLa cells were transfected with mPax8a (0.1 µg) and Grg4 (0.5 µg) expression plasmids and the luciferase gene TPO-luc (2.5 µg) and pRL-SV40 (10 ng). Δ_TPO-luc_ contains a mutated Pax8-binding site (Zannini et al., 1992). (G and H) Grg4-mediated repression of the tran scriptional activity of zebrafish zfPax2.1 and Drosophila dPax258. (I) Repression of the Pax5 transactivation function by Drosophila Groucho.
Fig. 8. The octapeptide motif of Pax5 mediates repression by Grg4. Transient transfection experiments with the expression plasmids indicated were performed in SP2/0 cells and subsequently evaluated as described in the legend to Figure 7.
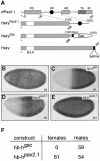
Fig. 9. The octapeptide motif functions as a repression domain in Drosophila embryos. (A) Schematic diagram of the Hairy derivatives used in the Sxl repression assay. The HairyPax2.1 protein was generated by fusing the Hairy sequences at amino acid 268 to a 90-amino-acid sequence (143–232) of the zebrafish Pax2.1 protein encompassing the conserved octapeptide motif (Pfeffer et al., 1998). The HairyGsc protein containing the eh1/GEH motif of Goosecoid (Gsc) was described previously (Jiménez et al., 1999). (B–E) Sxl repression by chimeric Hairy proteins in female blastoderm embryos of Drosophila. The HairyGsc (C) and HairyPax2.1 (D and E) proteins were expressed under the control of the hunchback (hb) promoter in wild-type (wt, C and D) or _gro_E48mat– mutant embryos (E). Sxl expression was analyzed in transgenic (C–E) and control (B) female embryos by whole-mount staining with a monoclonal antibody detecting the female-specific form of Sxl (Bopp et al., 1991). (F) Effects of HairyGsc and HairyPax2.1 expression on female viability. Wild-type females were crossed with hb transgenic males, and the number of viable progeny is indicated.
Fig. 10. Model of Grg4 recruitment by Pax5 (BSAP). (A) Summary of the identified interactions between Pax5 and Grg4. (B) Model ofGrg-mediated gene repression by Pax5. As different members of the Grg family are co-expressed with Pax5 throughout midbrain and B-cell development (Koop et al., 1996; D.Eberhard, unpublished data), we hypothesize that Pax5 can stably recruit Grg proteins to a specific promoter only in collaboration with a second Grg-binding transcription factor (X). CoA, coactivator protein(s); BTM, basal transcription machinery.
Similar articles
- Corecruitment of the Grg4 repressor by PU.1 is critical for Pax5-mediated repression of B-cell-specific genes.
Linderson Y, Eberhard D, Malin S, Johansson A, Busslinger M, Pettersson S. Linderson Y, et al. EMBO Rep. 2004 Mar;5(3):291-6. doi: 10.1038/sj.embor.7400089. Epub 2004 Feb 20. EMBO Rep. 2004. PMID: 14993928 Free PMC article. - C-terminal activating and inhibitory domains determine the transactivation potential of BSAP (Pax-5), Pax-2 and Pax-8.
Dörfler P, Busslinger M. Dörfler P, et al. EMBO J. 1996 Apr 15;15(8):1971-82. EMBO J. 1996. PMID: 8617244 Free PMC article. - FoxD3 and Grg4 physically interact to repress transcription and induce mesoderm in Xenopus.
Yaklichkin S, Steiner AB, Lu Q, Kessler DS. Yaklichkin S, et al. J Biol Chem. 2007 Jan 26;282(4):2548-57. doi: 10.1074/jbc.M607412200. Epub 2006 Nov 30. J Biol Chem. 2007. PMID: 17138566 Free PMC article. - Pax5 determines the identity of B cells from the beginning to the end of B-lymphopoiesis.
Nutt SL, Eberhard D, Horcher M, Rolink AG, Busslinger M. Nutt SL, et al. Int Rev Immunol. 2001 Feb;20(1):65-82. doi: 10.3109/08830180109056723. Int Rev Immunol. 2001. PMID: 11342298 Review. - Context-dependent regulation of Groucho/TLE-mediated repression.
Cinnamon E, Paroush Z. Cinnamon E, et al. Curr Opin Genet Dev. 2008 Oct;18(5):435-40. doi: 10.1016/j.gde.2008.07.010. Epub 2008 Sep 5. Curr Opin Genet Dev. 2008. PMID: 18721877 Review.
Cited by
- Roles of transducin-like enhancer of split (TLE) family proteins in tumorigenesis and immune regulation.
Yu G, Chen Y, Hu Y, Zhou Y, Ding X, Zhou X. Yu G, et al. Front Cell Dev Biol. 2022 Nov 11;10:1010639. doi: 10.3389/fcell.2022.1010639. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36438567 Free PMC article. Review. - Default repression and Notch signaling: Hairless acts as an adaptor to recruit the corepressors Groucho and dCtBP to Suppressor of Hairless.
Barolo S, Stone T, Bang AG, Posakony JW. Barolo S, et al. Genes Dev. 2002 Aug 1;16(15):1964-76. doi: 10.1101/gad.987402. Genes Dev. 2002. PMID: 12154126 Free PMC article. - The evolution of alternative splicing in the Pax family: the view from the Basal chordate amphioxus.
Short S, Holland LZ. Short S, et al. J Mol Evol. 2008 Jun;66(6):605-20. doi: 10.1007/s00239-008-9113-5. Epub 2008 May 14. J Mol Evol. 2008. PMID: 18473110 - Identification and characterization of Nasonia Pax genes.
Keller RG, Desplan C, Rosenberg MI. Keller RG, et al. Insect Mol Biol. 2010 Feb;19 Suppl 1(Suppl 1):109-20. doi: 10.1111/j.1365-2583.2009.00921.x. Insect Mol Biol. 2010. PMID: 20167022 Free PMC article. - From hematopoietic progenitors to B cells: mechanisms of lineage restriction and commitment.
Ramírez J, Lukin K, Hagman J. Ramírez J, et al. Curr Opin Immunol. 2010 Apr;22(2):177-84. doi: 10.1016/j.coi.2010.02.003. Epub 2010 Mar 6. Curr Opin Immunol. 2010. PMID: 20207529 Free PMC article. Review.
References
- Adams B., Dörfler,P., Aguzzi,A., Kozmik,Z., Urbánek,P., Maurer-Fogy,I. and Busslinger,M. (1992) Pax-5 encodes the transcription factor BSAP and is expressed in B lymphocytes, the developing CNS and adult testis. Genes Dev., 6, 1589–1607. - PubMed
- Alkema M.J., Jacobs,J., Voncken,J.W., Jenkins,N.A., Copeland,N.G., Satijn,D.P.E., Otte,A.P., Berns,A. and van Lohuizen,M. (1997) MPc2, a new murine homolog of the Drosophila Polycomb protein is a member of the mouse Polycomb transcriptional repressor complex. J. Mol. Biol., 273, 993–1003. - PubMed
- Bopp D., Bell,L.R., Cline,T.W. and Schedl,P. (1991) Developmental distribution of female-specific Sex-lethal proteins in Drosophila melanogaster. Genes Dev., 5, 403–415. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases