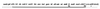An efficient recombination system for chromosome engineering in Escherichia coli - PubMed (original) (raw)
An efficient recombination system for chromosome engineering in Escherichia coli
D Yu et al. Proc Natl Acad Sci U S A. 2000.
Abstract
A recombination system has been developed for efficient chromosome engineering in Escherichia coli by using electroporated linear DNA. A defective lambda prophage supplies functions that protect and recombine an electroporated linear DNA substrate in the bacterial cell. The use of recombination eliminates the requirement for standard cloning as all novel joints are engineered by chemical synthesis in vitro and the linear DNA is efficiently recombined into place in vivo. The technology and manipulations required are simple and straightforward. A temperature-dependent repressor tightly controls prophage expression, and, thus, recombination functions can be transiently supplied by shifting cultures to 42 degrees C for 15 min. The efficient prophage recombination system does not require host RecA function and depends primarily on Exo, Beta, and Gam functions expressed from the defective lambda prophage. The defective prophage can be moved to other strains and can be easily removed from any strain. Gene disruptions and modifications of both the bacterial chromosome and bacterial plasmids are possible. This system will be especially useful for the engineering of large bacterial plasmids such as those from bacterial artificial chromosome libraries.
Figures
Figure 1
Description of the defective λ prophage on the E. coli chromosome. The defective prophage contains λ genes from_cI to int._ A deletion (dotted line) removes the right side of the prophage from cro through_attR_ and including bioA (21). On the chromosome, the nadA and gal operons are to the left of the prophage, and the bio genes without_bioA_ are to the right. Genes of the λ prophage are shown on the solid line, and genes of the host are shown on the broken line. _p_L and _p_R indicate the early left and right promoters of λ. attL and attR indicate the left and right attachment sites of λ. The λ genes and functions have been described (17).
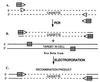
Figure 2
Strategy for generating recombinant DNA molecules and gene replacement. Three steps are outlined. (A) Recombinant oligonucleotides were chemically synthesized with the 5′ 30–50 nt (shaded rectangles) identical to sequences at the target and the 3′ 20 nt (arrowheads) homologous to the ends of the cassette to be introduced. A cassette is generated by PCR that is flanked by the 30- to 50-bp homologies present at the target. (B) Cells carrying the target DNA either on the chromosome or on a plasmid are induced for Exo, Beta, and Gam function. These cells are made competent for electroporation and mixed with the amplified cassette. (C) After electroporation, recombination occurs between the homologous sequences on the linear cassette and the target replacing the target segment with the cassette. The 50-nt_galK_ homology segments (rectangles) used for the experiment described in Table 2 are 5′GTTTGCGCGCAGTCAGCGATATCCATTTTCGCGAATCCGGAGTGTAAGAA and 5′'TTCATATTGTTCAGCGACAGCTTGCTGTACGGCAGGCACCAGCTCTTCCG.
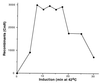
Figure 3
Effect of induction time on recombination. The strain DY330 was grown at 32°C to OD600 = 0.4–0.6, was induced at 42°C for the times indicated, and then was made electrocompetent. A linear_cat_ cassette (10 ng) was used to target the cIII kil gam genes of the prophage. Total CmR recombinants were plotted vs. the time of induction.
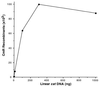
Figure 4
Effect of concentration of the linear DNA cassette on recombination. The strain DY330 was grown at 32°C to OD600 = 0.4–0.6, was induced at 42°C for 15 min, and then was made electrocompetent. Different amounts (1, 10, 100, 300, and 1,000 ng) of a linear cat cassette (1 kbp in length) were used to target the cIII kil gam genes of the prophage. Total CmR recombinants were plotted vs. the DNA amount at 42°C.
Figure 5
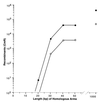
Effect of homologous arm length on recombination. The strains DY330 (filled circles) and DY331 (open circles) were grown at 32°C, were induced at 42°C for 15 min, and then were made relectrocompetent. A linear cat cassette (100 ng) was used to target the_cIII kil gam_ genes of the prophage. The homologous arm length of the cassette was varied from 0 to 1,000 bp. The primers containing the 0- to 50-bp homologies were chemically synthesized as described (Fig. 2). The cassette containing 1,000-bp homologous arms was made by PCR using primers 1,000 bp away on each side of an existing (cIII kil gam)<>cat disruption in the cell. Total CmR recombinants were plotted vs. the homologous arm length.
Similar articles
- A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA.
Lee EC, Yu D, Martinez de Velasco J, Tessarollo L, Swing DA, Court DL, Jenkins NA, Copeland NG. Lee EC, et al. Genomics. 2001 Apr 1;73(1):56-65. doi: 10.1006/geno.2000.6451. Genomics. 2001. PMID: 11352566 - Recombineering with overlapping single-stranded DNA oligonucleotides: testing a recombination intermediate.
Yu D, Sawitzke JA, Ellis H, Court DL. Yu D, et al. Proc Natl Acad Sci U S A. 2003 Jun 10;100(12):7207-12. doi: 10.1073/pnas.1232375100. Epub 2003 May 27. Proc Natl Acad Sci U S A. 2003. PMID: 12771385 Free PMC article. - Bacterial artificial chromosome engineering.
Swaminathan S, Sharan SK. Swaminathan S, et al. Methods Mol Biol. 2004;256:89-106. doi: 10.1385/1-59259-753-X:089. Methods Mol Biol. 2004. PMID: 15024162 No abstract available. - [Red/ET recombination and its biomedical applications].
Wang JP, Zhang YM. Wang JP, et al. Sheng Wu Gong Cheng Xue Bao. 2005 May;21(3):502-6. Sheng Wu Gong Cheng Xue Bao. 2005. PMID: 16108384 Review. Chinese. - [Recombineering and its application].
Zhou JG, Hong X, Huang CF. Zhou JG, et al. Yi Chuan Xue Bao. 2003 Oct;30(10):983-8. Yi Chuan Xue Bao. 2003. PMID: 14669518 Review. Chinese.
Cited by
- The design and engineering of synthetic genomes.
James JS, Dai J, Chew WL, Cai Y. James JS, et al. Nat Rev Genet. 2024 Nov 6. doi: 10.1038/s41576-024-00786-y. Online ahead of print. Nat Rev Genet. 2024. PMID: 39506144 Review. - Reducing competition between msd and genomic DNA improves retron editing efficiency.
Ni Y, Wang Y, Shi X, Yu F, Ruan Q, Tian N, He J, Wang X. Ni Y, et al. EMBO Rep. 2024 Nov 5. doi: 10.1038/s44319-024-00311-6. Online ahead of print. EMBO Rep. 2024. PMID: 39501049 - Diverse anti-defence systems are encoded in the leading region of plasmids.
Samuel B, Mittelman K, Croitoru SY, Ben Haim M, Burstein D. Samuel B, et al. Nature. 2024 Nov;635(8037):186-192. doi: 10.1038/s41586-024-07994-w. Epub 2024 Oct 9. Nature. 2024. PMID: 39385022 Free PMC article. - Diribonuclease activity eliminates toxic diribonucleotide accumulation.
Kim SK, Orr MW, Turdiev H, Jenkins CC, Lormand JD, Myers TM, Burnim AA, Carter JA, Kung WC, Jiang X, Sondermann H, Winkler WC, Lee VT. Kim SK, et al. Cell Rep. 2024 Sep 24;43(9):114759. doi: 10.1016/j.celrep.2024.114759. Epub 2024 Sep 13. Cell Rep. 2024. PMID: 39276351 Free PMC article. - A review of HSV pathogenesis, vaccine development, and advanced applications.
Bai L, Xu J, Zeng L, Zhang L, Zhou F. Bai L, et al. Mol Biomed. 2024 Aug 29;5(1):35. doi: 10.1186/s43556-024-00199-7. Mol Biomed. 2024. PMID: 39207577 Free PMC article. Review.
References
- Szostak J W, Orr-Weaver T L, Rothstein R J, Stahl F W. Cell. 1983;33:25–35. - PubMed
- Yamamoto T, Moerschell R P, Wakem L P, Ferguson D, Sherman F. Yeast. 1992;8:935–948. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources