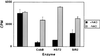The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases - PubMed (original) (raw)
The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases
J Landry et al. Proc Natl Acad Sci U S A. 2000.
Abstract
Homologs of the chromatin-bound yeast silent information regulator 2 (SIR2) protein are found in organisms from all biological kingdoms. SIR2 itself was originally discovered to influence mating-type control in haploid cells by locus-specific transcriptional silencing. Since then, SIR2 and its homologs have been suggested to play additional roles in suppression of recombination, chromosomal stability, metabolic regulation, meiosis, and aging. Considering the far-ranging nature of these functions, a major experimental goal has been to understand the molecular mechanism(s) by which this family of proteins acts. We report here that members of the SIR2 family catalyze an NAD-nicotinamide exchange reaction that requires the presence of acetylated lysines such as those found in the N termini of histones. Significantly, these enzymes also catalyze histone deacetylation in a reaction that absolutely requires NAD, thereby distinguishing them from previously characterized deacetylases. The enzymes are active on histone substrates that have been acetylated by both chromatin assembly-linked and transcription-related acetyltransferases. Contrary to a recent report, we find no evidence that these proteins ADP-ribosylate histones. Discovery of an intrinsic deacetylation activity for the conserved SIR2 family provides a mechanism for modifying histones and other proteins to regulate transcription and diverse biological processes.
Figures
Figure 1
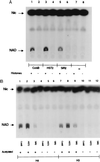
NAD–nicotinamide exchange reactions with CobB, HST2, or SIR2. (A) Reactions were performed with (lanes 1, 3, 5, and 7) and without (lanes 2, 4, 6, and 8) histones, as indicated. (B) Reactions with acetylated (lanes 1–3, 7–9) or unacetylated (lanes 4–6, 10–12) H4 or H3 peptides. Peptides were acetylated as described in Materials and Methods.
Figure 2
Histone deacetylation by CobB, HST2, and SIR2 depends on NAD. The cpm present in the histones after enzyme incubation are depicted. Error bars show the variation between two independent experiments.
Figure 3
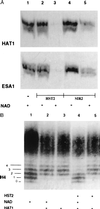
Histone deacetylation analyzed by gel electrophoresis. (A) An SDS/PAGE gel of histones acetylated with [3H]acetyl-CoA by HAT1 or by ESA1, as indicated, and then treated with no enzyme (lane 1), with HST2 (lanes 2 and 3), or with SIR2 (lanes 4 and 5). A fluorogram of the gel is depicted. (B) A Triton–acid–urea gel of chicken erythrocyte histones acetylated with HAT1 followed by deacetylation by HST2. The gel was stained with Coomassie blue to visualize the histone isoforms. The numbers 0–4 on the left refer to isoforms of H4 differing in charge, with 0 corresponding to the most positively charged isoform and 4 corresponding to the least positively charged form. Lane 1, histones incubated without HAT1; lanes 2 and 3, histones acetylated with HAT1, in the presence or absence of NAD; lanes 4 and 5, histones acetylated by HAT1, incubated with HST2, with and without NAD.
Figure 4
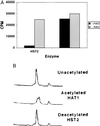
Deacetylation of an H4 peptide by HST2. The peptide was initially acetylated by HAT1. (A) H4 peptide deacetylation by HST2 depends on NAD. The cpm present in the peptide after enzyme incubation are depicted. (B) HPLC traces of the input unacetylated H4 peptide, the peptide after acetylation by HAT1, and the peptide after subsequent deacetylation by HST2.
Similar articles
- Coupling of histone deacetylation to NAD breakdown by the yeast silencing protein Sir2: Evidence for acetyl transfer from substrate to an NAD breakdown product.
Tanny JC, Moazed D. Tanny JC, et al. Proc Natl Acad Sci U S A. 2001 Jan 16;98(2):415-20. doi: 10.1073/pnas.98.2.415. Epub 2000 Dec 26. Proc Natl Acad Sci U S A. 2001. PMID: 11134535 Free PMC article. - Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase.
Imai S, Armstrong CM, Kaeberlein M, Guarente L. Imai S, et al. Nature. 2000 Feb 17;403(6771):795-800. doi: 10.1038/35001622. Nature. 2000. PMID: 10693811 - SIR2: the biochemical mechanism of NAD(+)-dependent protein deacetylation and ADP-ribosyl enzyme intermediates.
Sauve AA, Schramm VL. Sauve AA, et al. Curr Med Chem. 2004 Apr;11(7):807-26. doi: 10.2174/0929867043455675. Curr Med Chem. 2004. PMID: 15078167 Review. - A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family.
Smith JS, Brachmann CB, Celic I, Kenna MA, Muhammad S, Starai VJ, Avalos JL, Escalante-Semerena JC, Grubmeyer C, Wolberger C, Boeke JD. Smith JS, et al. Proc Natl Acad Sci U S A. 2000 Jun 6;97(12):6658-63. doi: 10.1073/pnas.97.12.6658. Proc Natl Acad Sci U S A. 2000. PMID: 10841563 Free PMC article. - The Sir2 protein family: A novel deacetylase for gene silencing and more.
Shore D. Shore D. Proc Natl Acad Sci U S A. 2000 Dec 19;97(26):14030-2. doi: 10.1073/pnas.011506198. Proc Natl Acad Sci U S A. 2000. PMID: 11114164 Free PMC article. Review. No abstract available.
Cited by
- Biotinylation of lysine method identifies acetylated histone H3 lysine 79 in Saccharomyces cerevisiae as a substrate for Sir2.
Bheda P, Swatkoski S, Fiedler KL, Boeke JD, Cotter RJ, Wolberger C. Bheda P, et al. Proc Natl Acad Sci U S A. 2012 Apr 17;109(16):E916-25. doi: 10.1073/pnas.1121471109. Epub 2012 Apr 2. Proc Natl Acad Sci U S A. 2012. PMID: 22474337 Free PMC article. - A family of tetraspans organizes cargo for sorting into multivesicular bodies.
MacDonald C, Payne JA, Aboian M, Smith W, Katzmann DJ, Piper RC. MacDonald C, et al. Dev Cell. 2015 May 4;33(3):328-42. doi: 10.1016/j.devcel.2015.03.007. Dev Cell. 2015. PMID: 25942624 Free PMC article. - Genome-wide analysis of functional sirtuin chromatin targets in yeast.
Li M, Valsakumar V, Poorey K, Bekiranov S, Smith JS. Li M, et al. Genome Biol. 2013 May 27;14(5):R48. doi: 10.1186/gb-2013-14-5-r48. Genome Biol. 2013. PMID: 23710766 Free PMC article. - Structural basis for nicotinamide inhibition and base exchange in Sir2 enzymes.
Sanders BD, Zhao K, Slama JT, Marmorstein R. Sanders BD, et al. Mol Cell. 2007 Feb 9;25(3):463-72. doi: 10.1016/j.molcel.2006.12.022. Mol Cell. 2007. PMID: 17289592 Free PMC article. - Sir3-dependent assembly of supramolecular chromatin structures in vitro.
Georgel PT, Palacios DeBeer MA, Pietz G, Fox CA, Hansen JC. Georgel PT, et al. Proc Natl Acad Sci U S A. 2001 Jul 17;98(15):8584-9. doi: 10.1073/pnas.151258798. Epub 2001 Jul 10. Proc Natl Acad Sci U S A. 2001. PMID: 11447281 Free PMC article.
References
- Loo S, Rine J. Annu Rev Cell Dev Biol. 1995;11:519–548. - PubMed
- Sherman J M, Pillus L. Trends Genet. 1997;13:308–313. - PubMed
- Guarente L. Nat Genet. 1999;23:281–285. - PubMed
- Hecht A, Laroche T, Strahl-Bolsinger S, Gasser S M, Grunstein M. Cell. 1995;80:583–592. - PubMed
- Straight A F, Shou W, Dowd G J, Turck C W, Deshaies R J, Johnson A D, Moazed D. Cell. 1999;97:245–256. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM054778/GM/NIGMS NIH HHS/United States
- R56 GM028220/GM/NIGMS NIH HHS/United States
- R01 GM055641/GM/NIGMS NIH HHS/United States
- GM55641/GM/NIGMS NIH HHS/United States
- GM54778/GM/NIGMS NIH HHS/United States
- R01 GM028220/GM/NIGMS NIH HHS/United States
- GM28220/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases