Expression of hepatitis C virus proteins interferes with the antiviral action of interferon independently of PKR-mediated control of protein synthesis - PubMed (original) (raw)
Expression of hepatitis C virus proteins interferes with the antiviral action of interferon independently of PKR-mediated control of protein synthesis
C François et al. J Virol. 2000 Jun.
Abstract
Hepatitis C virus (HCV) of genotype 1 is the most resistant to interferon (IFN) therapy. Here, we have analyzed the response to IFN of the human cell line UHCV-11 engineered to inducibly express the entire HCV genotype 1a polyprotein. IFN-treated, induced UHCV cells were found to better support the growth of encephalomyocarditis virus (EMCV) than IFN-treated, uninduced cells. This showed that expression of the HCV proteins allowed the development of a partial resistance to the antiviral action of IFN. The nonstructural 5A (NS5A) protein of HCV has been reported to inhibit PKR, an IFN-induced kinase involved in the antiviral action of IFN, at the level of control of protein synthesis through the phosphorylation of the initiation factor eIF2alpha (M. Gale, Jr., C. M. Blakely, B. Kwieciszewski, S. L. Tan, M. Dossett, N. M. Tang, M. J. Korth, S. J. Polyak, D. R. Gretch, and M. G. Katze, Mol. Cell. Biol. 18:5208-5218, 1998). Accordingly, cell lines inducibly expressing NS5A were found to rescue EMCV growth (S. J. Polyak, D. M. Paschal, S. McArdle, M. J. Gale, Jr., D. Moradpour, and D. R. Gretch, Hepatology 29:1262-1271, 1999). In the present study we analyzed whether the resistance of UHCV-11 cells to IFN could also be attributed to inhibition of PKR. Confocal laser scanning microscopy showed no colocalization of PKR, which is diffuse throughout the cytoplasm, and the induced HCV proteins, which localize around the nucleus within the endoplasmic reticulum. The effect of expression of HCV proteins on PKR activity was assayed in a reporter assay and by direct analysis of the in vivo phosphorylation of eIF2alpha after treatment of cells with poly(I)-poly(C). We found that neither PKR activity nor eIF2alpha phosphorylation was affected by coexpression of the HCV proteins. In conclusion, expression of HCV proteins in their biological context interferes with the development of the antiviral action of IFN. Although the possibility that some inhibition of PKR (by either NS5A or another viral protein) occurs at a very localized level cannot be excluded, the resistance to IFN, resulting from the expression of the HCV proteins, cannot be explained solely by inhibition of the negative control of translation by PKR.
Figures
FIG. 1
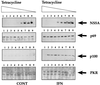
Regulation of HCV NS5A protein expression in UHCV-11 cells and response to IFN with respect to PKR and 2-5A synthetase induction. UHCV-11 cells were seeded in four six-well tissue culture plates at a density of 300,000/well in the presence of tetracycline. After 24 h, they were washed three times with PBS to remove tetracycline and then incubated in culture medium containing different concentrations of tetracycline (1,000, 500, 100, 50, 10, 5, 1, 0.5, or 0 ng/ml [from left to right, lanes 1 to 9]). One set of cells was incubated in the absence of IFN (control [CONT]), and the other set was incubated in the presence of IFN at 500 U/ml. After 18 h of incubation, the cells were lysed in 300 μl of low-salt buffer I (containing 40 mM of NaCl instead of 400 mM) as described in Materials and Methods. The proteins contained in 50 μl of each extract (equivalent to 100,000 cells) were separated by SDS–12.5% PAGE and analyzed by immunoblotting for the presence of NS5A, p69 2-5A synthetase, p100 2-5A synthetase, and PKR.
FIG. 2
Analysis of in vitro activity of PKR and 2-5A synthetases from UHCV-11 cells. (A) PKR activity. Extracts of UHCV-11 cells that had (Tet−) or had not (Tet+) been induced to express the polyprotein and that had or had not (control [CONT]) been treated with IFN were prepared in buffer I. Immunoprecipitation of PKR and an in vitro phosphorylation assay were performed with extracts corresponding to 107 cells as described in Materials and Methods. PKR activity was assayed in the absence (0) and in the presence of poly(I)-poly(C) (IC) at 0.5 or 1 μg/ml or in the presence of heparin (H) at 10 U/ml (used as activators). Fifteen minutes after the beginning of the phosphorylation reaction, 2 μl of a purified preparation of eIF2 was added to each of the samples containing no activator or containing heparin (asterisks), and the reactions were continued for another 15 min. The reactions were stopped by addition of an equal amount of 2× SDS sample buffer, and the proteins were separated by SDS–12.5% PAGE. (B) Immunoblotting for PKR and NS5A. Ten percent of each of the crude extracts used for the immunoprecipitation detailed above and corresponding to untreated and IFN-treated UHCV cells that had or had not been induced to produce the HCV polyprotein were analyzed by immunoblotting for induction of PKR after IFN treatment (immunoblot PKR) and for induction of NS5A by tetracycline removal (immunoblot NS5A). (C) 2-5A synthetase activity. Extracts from UHCV-11 cells repressed (TET+) or induced (TET−) for the expression of the polyprotein and extracts from Daudi cells, used as controls, were immunoprecipitated with antibodies against either the p69 or the p100 form of 2-5A synthetase and analyzed for their capacity to synthesize 2-5A oligomers as described in Materials and Methods.
FIG. 2
Analysis of in vitro activity of PKR and 2-5A synthetases from UHCV-11 cells. (A) PKR activity. Extracts of UHCV-11 cells that had (Tet−) or had not (Tet+) been induced to express the polyprotein and that had or had not (control [CONT]) been treated with IFN were prepared in buffer I. Immunoprecipitation of PKR and an in vitro phosphorylation assay were performed with extracts corresponding to 107 cells as described in Materials and Methods. PKR activity was assayed in the absence (0) and in the presence of poly(I)-poly(C) (IC) at 0.5 or 1 μg/ml or in the presence of heparin (H) at 10 U/ml (used as activators). Fifteen minutes after the beginning of the phosphorylation reaction, 2 μl of a purified preparation of eIF2 was added to each of the samples containing no activator or containing heparin (asterisks), and the reactions were continued for another 15 min. The reactions were stopped by addition of an equal amount of 2× SDS sample buffer, and the proteins were separated by SDS–12.5% PAGE. (B) Immunoblotting for PKR and NS5A. Ten percent of each of the crude extracts used for the immunoprecipitation detailed above and corresponding to untreated and IFN-treated UHCV cells that had or had not been induced to produce the HCV polyprotein were analyzed by immunoblotting for induction of PKR after IFN treatment (immunoblot PKR) and for induction of NS5A by tetracycline removal (immunoblot NS5A). (C) 2-5A synthetase activity. Extracts from UHCV-11 cells repressed (TET+) or induced (TET−) for the expression of the polyprotein and extracts from Daudi cells, used as controls, were immunoprecipitated with antibodies against either the p69 or the p100 form of 2-5A synthetase and analyzed for their capacity to synthesize 2-5A oligomers as described in Materials and Methods.
FIG. 2
Analysis of in vitro activity of PKR and 2-5A synthetases from UHCV-11 cells. (A) PKR activity. Extracts of UHCV-11 cells that had (Tet−) or had not (Tet+) been induced to express the polyprotein and that had or had not (control [CONT]) been treated with IFN were prepared in buffer I. Immunoprecipitation of PKR and an in vitro phosphorylation assay were performed with extracts corresponding to 107 cells as described in Materials and Methods. PKR activity was assayed in the absence (0) and in the presence of poly(I)-poly(C) (IC) at 0.5 or 1 μg/ml or in the presence of heparin (H) at 10 U/ml (used as activators). Fifteen minutes after the beginning of the phosphorylation reaction, 2 μl of a purified preparation of eIF2 was added to each of the samples containing no activator or containing heparin (asterisks), and the reactions were continued for another 15 min. The reactions were stopped by addition of an equal amount of 2× SDS sample buffer, and the proteins were separated by SDS–12.5% PAGE. (B) Immunoblotting for PKR and NS5A. Ten percent of each of the crude extracts used for the immunoprecipitation detailed above and corresponding to untreated and IFN-treated UHCV cells that had or had not been induced to produce the HCV polyprotein were analyzed by immunoblotting for induction of PKR after IFN treatment (immunoblot PKR) and for induction of NS5A by tetracycline removal (immunoblot NS5A). (C) 2-5A synthetase activity. Extracts from UHCV-11 cells repressed (TET+) or induced (TET−) for the expression of the polyprotein and extracts from Daudi cells, used as controls, were immunoprecipitated with antibodies against either the p69 or the p100 form of 2-5A synthetase and analyzed for their capacity to synthesize 2-5A oligomers as described in Materials and Methods.
FIG. 3
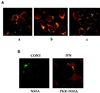
Subcellular localization of PKR, 2-5A synthetase, and NS5A in UHCV-11 cells. (A) UHCV-11 cells, seeded in eight-well chamber slides (Lab-Tek), were induced for the full expression of the HCV polyprotein and analyzed for the colocalization of NS5A (green fluorescence) and either PKR (a), p69 2-5A synthetase (b), or p100 2-5A synthetase (c) (red fluorescence). (B) UHCV-11 cells, seeded in eight-well chamber slides (Lab-Tek), were induced for the full expression of the HCV polyprotein in the absence (control [CONT]) or presence of IFN of and analyzed for the effect of IFN on localization of NS5A (green fluorescence) and PKR (red fluorescence).
FIG. 4
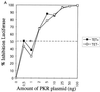
Expression of HCV polyprotein does not reverse the PKR-mediated inhibition of protein synthesis, whereas TRBP does. (A) UHCV-11 cells were transfected by a microtransfection technique described in Materials and Methods. To each well (20,000 cells) was added 100 ng of pHIV1 LTR-L and 0.5 to 100 ng of pcDNA1/Amp (PKR) as indicated. All samples were adjusted to contain the same amount of DNA by addition of the empty pcDNA1/Amp vector. The transfection assay was performed with cells either repressed (TET+) (closed squares) or induced (TET−) (open squares) for the expression of the HCV proteins. The results are expressed as the percentage of inhibition of reporter expression when transfection took place in the presence of PKR compared with its transfection in the absence of the PKR plasmid. (B and C) UHCV-11 cells either repressed (B) or induced (C) for the expression of the polyprotein were transfected with pHIV1 LTR-L and increasing concentrations of pcDNA1/Amp (PKR), either alone (closed squares) as for panel A or in the presence of 300 ng of pcDNA1/Amp (TRBP2) (open circles). The results are expressed as the percentage of inhibition of reporter expression by PKR. Addition of TRBP to the cells allows a shift of the ID50 of PKR from the 0.8- to 1-ng range to the 8- to 10-ng range, thus indicating its ability to reverse the PKR-mediated inhibition of protein synthesis. In each graph, the ID50 of PKR is represented by an asterisk placed where the graph crosses the 50% inhibition value (broken line).
FIG. 4
Expression of HCV polyprotein does not reverse the PKR-mediated inhibition of protein synthesis, whereas TRBP does. (A) UHCV-11 cells were transfected by a microtransfection technique described in Materials and Methods. To each well (20,000 cells) was added 100 ng of pHIV1 LTR-L and 0.5 to 100 ng of pcDNA1/Amp (PKR) as indicated. All samples were adjusted to contain the same amount of DNA by addition of the empty pcDNA1/Amp vector. The transfection assay was performed with cells either repressed (TET+) (closed squares) or induced (TET−) (open squares) for the expression of the HCV proteins. The results are expressed as the percentage of inhibition of reporter expression when transfection took place in the presence of PKR compared with its transfection in the absence of the PKR plasmid. (B and C) UHCV-11 cells either repressed (B) or induced (C) for the expression of the polyprotein were transfected with pHIV1 LTR-L and increasing concentrations of pcDNA1/Amp (PKR), either alone (closed squares) as for panel A or in the presence of 300 ng of pcDNA1/Amp (TRBP2) (open circles). The results are expressed as the percentage of inhibition of reporter expression by PKR. Addition of TRBP to the cells allows a shift of the ID50 of PKR from the 0.8- to 1-ng range to the 8- to 10-ng range, thus indicating its ability to reverse the PKR-mediated inhibition of protein synthesis. In each graph, the ID50 of PKR is represented by an asterisk placed where the graph crosses the 50% inhibition value (broken line).
FIG. 4
Expression of HCV polyprotein does not reverse the PKR-mediated inhibition of protein synthesis, whereas TRBP does. (A) UHCV-11 cells were transfected by a microtransfection technique described in Materials and Methods. To each well (20,000 cells) was added 100 ng of pHIV1 LTR-L and 0.5 to 100 ng of pcDNA1/Amp (PKR) as indicated. All samples were adjusted to contain the same amount of DNA by addition of the empty pcDNA1/Amp vector. The transfection assay was performed with cells either repressed (TET+) (closed squares) or induced (TET−) (open squares) for the expression of the HCV proteins. The results are expressed as the percentage of inhibition of reporter expression when transfection took place in the presence of PKR compared with its transfection in the absence of the PKR plasmid. (B and C) UHCV-11 cells either repressed (B) or induced (C) for the expression of the polyprotein were transfected with pHIV1 LTR-L and increasing concentrations of pcDNA1/Amp (PKR), either alone (closed squares) as for panel A or in the presence of 300 ng of pcDNA1/Amp (TRBP2) (open circles). The results are expressed as the percentage of inhibition of reporter expression by PKR. Addition of TRBP to the cells allows a shift of the ID50 of PKR from the 0.8- to 1-ng range to the 8- to 10-ng range, thus indicating its ability to reverse the PKR-mediated inhibition of protein synthesis. In each graph, the ID50 of PKR is represented by an asterisk placed where the graph crosses the 50% inhibition value (broken line).
FIG. 5
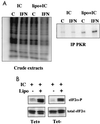
Poly(I)-poly(C)-induced phosphorylation of eIF2α is not affected by induction of the HCV proteins. (A) UHCV-11 cells were seeded at a density of 106/10-cm-diameter petri dish. After 24 h, they were washed three times with PBS to remove tetracycline and then incubated in culture medium alone (control [C]) or containing IFN at 500 U/ml. After 18 h of incubation, the cells were washed twice in phosphate-free, serum-free medium and further incubated in 2.5 ml of this medium supplemented with 100 μg of poly(I)-poly(C) (PL Biochemicals), either alone (IC) or mixed with 25 μg of Lipofectin (Gibco BRL) (lipo+IC). [32P]orthophosphate (Amersham) was then added (750 μCi/dish), and an incubation was carried out for 90 min at 37°C. The medium was removed, the cells were washed twice and scraped off in PBS, and the cell pellets were recovered by centrifugation and lysed with 600 μl of buffer I supplemented with 10 mM β-glycerophosphate, 10 mM NaF, 10 mM _p_-nitrophenyl-phosphate, and 300 μM Na3VO4 as phosphatase inhibitors. After centrifugation at 12,000 × g, 5 μl of each of the crude extracts was subjected to SDS–12.5% PAGE and the rest was immunoprecipitated with anti-PKR antibodies as described in Materials and Methods. After incubation at 4°C for 18 h, the beads were washed three times with buffer I and the proteins were separated by SDS–12.5% PAGE. IPLab Gel-based quantification gave estimates of 1.12-fold [treatment with poly(I)-poly(C) alone] and 3.2-fold [treatment with Lipofectin plus poly(I)-poly(C)] for the increases in intensity of the phosphorylated bands in the IFN lane compared with the control lanes. (B) UHCV-11 cells were treated with poly(I)-poly(C) either alone or in the presence of Lipofectin (Lipo) as for the experiment shown in A, except that all dishes of cells had been previously treated with IFN and induced (Tet−) or not induced (Tet+) for expression of the HCV proteins. The proteins of the cell extracts were separated by SDS–12.5% PAGE and analyzed by immunoblotting. Use of antibodies directed specifically against a phosphorylated eIF2α peptide (eIF2α-P) demonstrated that eIF2α phosphorylation had occurred, whereas antibodies directed against total eIF2 (eIF2α) revealed the total levels of eIF2α. In this experiment, eIF2α was found to migrate as two bands in the gel. Both bands were specific since they were recognized by the two different anti-eIF2α antibodies.
Similar articles
- Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.
Salisbury L, Baraitser L. Salisbury L, et al. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review. - Antioxidants for female subfertility.
Showell MG, Mackenzie-Proctor R, Jordan V, Hart RJ. Showell MG, et al. Cochrane Database Syst Rev. 2020 Aug 27;8(8):CD007807. doi: 10.1002/14651858.CD007807.pub4. Cochrane Database Syst Rev. 2020. PMID: 32851663 Free PMC article. - Comparison of Two Modern Survival Prediction Tools, SORG-MLA and METSSS, in Patients With Symptomatic Long-bone Metastases Who Underwent Local Treatment With Surgery Followed by Radiotherapy and With Radiotherapy Alone.
Lee CC, Chen CW, Yen HK, Lin YP, Lai CY, Wang JL, Groot OQ, Janssen SJ, Schwab JH, Hsu FM, Lin WH. Lee CC, et al. Clin Orthop Relat Res. 2024 Dec 1;482(12):2193-2208. doi: 10.1097/CORR.0000000000003185. Epub 2024 Jul 23. Clin Orthop Relat Res. 2024. PMID: 39051924 - Falls prevention interventions for community-dwelling older adults: systematic review and meta-analysis of benefits, harms, and patient values and preferences.
Pillay J, Gaudet LA, Saba S, Vandermeer B, Ashiq AR, Wingert A, Hartling L. Pillay J, et al. Syst Rev. 2024 Nov 26;13(1):289. doi: 10.1186/s13643-024-02681-3. Syst Rev. 2024. PMID: 39593159 Free PMC article. - Antioxidants for female subfertility.
Showell MG, Mackenzie-Proctor R, Jordan V, Hart RJ. Showell MG, et al. Cochrane Database Syst Rev. 2017 Jul 28;7(7):CD007807. doi: 10.1002/14651858.CD007807.pub3. Cochrane Database Syst Rev. 2017. PMID: 28752910 Free PMC article. Updated. Review.
Cited by
- Effects of length and location on the cellular response to double-stranded RNA.
Wang Q, Carmichael GG. Wang Q, et al. Microbiol Mol Biol Rev. 2004 Sep;68(3):432-52, table of contents. doi: 10.1128/MMBR.68.3.432-452.2004. Microbiol Mol Biol Rev. 2004. PMID: 15353564 Free PMC article. Review. - Involvement of PKR and RNase L in translational control and induction of apoptosis after Hepatitis C polyprotein expression from a vaccinia virus recombinant.
Gómez CE, Vandermeeren AM, García MA, Domingo-Gil E, Esteban M. Gómez CE, et al. Virol J. 2005 Sep 12;2:81. doi: 10.1186/1743-422X-2-81. Virol J. 2005. PMID: 16156900 Free PMC article. - Hepatitis C virus nonstructural 5A protein and interferon resistance: a new model for testing the reliability of mutational analyses.
Sarrazin C, Herrmann E, Bruch K, Zeuzem S. Sarrazin C, et al. J Virol. 2002 Nov;76(21):11079-90. doi: 10.1128/jvi.76.21.11079-11090.2002. J Virol. 2002. PMID: 12368350 Free PMC article. - Tricks and threats of RNA viruses - towards understanding the fate of viral RNA.
Markiewicz L, Drazkowska K, Sikorski PJ. Markiewicz L, et al. RNA Biol. 2021 May;18(5):669-687. doi: 10.1080/15476286.2021.1875680. Epub 2021 Feb 22. RNA Biol. 2021. PMID: 33618611 Free PMC article. Review. - Hepatitis C virus comes for dinner: How the hepatitis C virus interferes with autophagy.
Ploen D, Hildt E. Ploen D, et al. World J Gastroenterol. 2015 Jul 28;21(28):8492-507. doi: 10.3748/wjg.v21.i28.8492. World J Gastroenterol. 2015. PMID: 26229393 Free PMC article. Review.
References
- Anderson S L, Carton J M, Lou J, Xing L, Rubin B Y. Interferon-induced guanylate binding protein-1 (GBP-1) mediates an antiviral effect against vesicular stomatitis virus and encephalomyocarditis virus. Virology. 1999;256:8–14. - PubMed
- Barber G N, Jagus R, Meurs E F, Hovanessian A G, Katze M G. Molecular mechanisms responsible for malignant transformation by regulatory and catalytic domain variants of the interferon-induced enzyme RNA-dependent protein kinase. J Biol Chem. 1995;270:17423–17428. - PubMed
- Besse S, Rebouillat D, Marie I, Puvion-Dutilleul F, Hovanessian A G. Ultrastructural localization of interferon-inducible double-stranded RNA-activated enzymes in human cells. Exp Cell Res. 1998;239:379–392. - PubMed
- Chebath J, Benech P, Revel M, Vigneron M. Constitutive expression of (2′-5′) oligo A synthetase confers resistance to picornavirus infection. Nature. 1987;330:587–588. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources