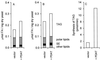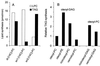Phospholipid:diacylglycerol acyltransferase: an enzyme that catalyzes the acyl-CoA-independent formation of triacylglycerol in yeast and plants - PubMed (original) (raw)
Phospholipid:diacylglycerol acyltransferase: an enzyme that catalyzes the acyl-CoA-independent formation of triacylglycerol in yeast and plants
A Dahlqvist et al. Proc Natl Acad Sci U S A. 2000.
Abstract
Triacylglycerol (TAG) is known to be synthesized in a reaction that uses acyl-CoA as acyl donor and diacylglycerol (DAG) as acceptor, and which is catalyzed by the enzyme acyl-CoA:diacylglycerol acyltransferase. We have found that some plants and yeast also have an acyl-CoA-independent mechanism for TAG synthesis, which uses phospholipids as acyl donors and DAG as acceptor. This reaction is catalyzed by an enzyme that we call phospholipid:diacylglycerol acyltransferase, or PDAT. PDAT was characterized in microsomal preparations from three different oil seeds: sunflower, castor bean, and Crepis palaestina. We found that the specificity of the enzyme for the acyl group in the phospholipid varies between these species. Thus, C. palaestina PDAT preferentially incorporates vernoloyl groups into TAG, whereas PDAT from castor bean incorporates both ricinoleoyl and vernoloyl groups. We further found that PDAT activity also is present in yeast microsomes. The substrate specificity of this PDAT depends on the head group of the acyl donor, the acyl group transferred, and the acyl chains of the acceptor DAG. The gene encoding the enzyme was identified. The encoded PDAT protein is related to lecithin:cholesterol acyltransferase, which catalyzes the acyl-CoA-independent synthesis of cholesterol esters. However, budding yeast PDAT and its relatives in fission yeast and Arabidopsis form a distinct branch within this protein superfamily, indicating that a separate PDAT enzyme arose at an early point in evolution.
Figures
Figure 1
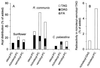
Metabolism of 14C-labeled PC into the neutral lipid fraction by plant microsomes. (A) Microsomes from developing seeds of sunflower, R. communis, and C. palaestina were incubated for 80 min at 30°C with PC (8 nmol) having oleic acid in its _sn_-1 position, and 14C-labeled oleic, ricinoleic, or vernolic acid in its _sn_-2 position. Radioactivity incorporated into TAG (open bars), DAG (filled bars), and unesterified fatty acids (hatched bars) is shown as percentage of added labeled substrate. (B) Synthesis in vitro of TAG carrying two vernoloyl and one [14C]ricinoleoyl group by microsomes from R. communis. The substrates added were unlabeled divernoloyl-DAG (5 nmol), together with either _sn_-1-oleoyl-_sn_-2-[14C]ricinoleoyl-DAG (0.4 nmol, 7,700 dpm/nmol) or _sn_-1-oleoyl-_sn_-2-[14C]ricinoleoyl-PC (0.4 nmol, 7,700 dpm/nmol). The microsomes were incubated with the substrates for 30 min at 30°C as described in Materials and Methods. The data shown are the average of two experiments.
Figure 2
PDAT activity in yeast microsomes, as visualized by autoradiogram of neutral lipid products separated on TLC. Microsomal membranes from the wild-type yeast strain FY1679 (lanes 1–3), a congenic yeast strain [FVKT004–04C(AL)] that is disrupted for YNR008w (lane 4), or the same disruption strain transformed with the plasmid pUS1, containing the YNR008w gene behind its native promoter (lane 5), were assayed for PDAT activity. As substrates, we used 2 nmol of _sn_-1-oleoyl-_sn_-2-[14C]ricinoleoyl-PC together with either 5 nmol of dioleoyl-DAG (lanes 2, 4, and 5) or _rac_-oleoyl-vernoloyl-DAG (lane 3). The enzymatic assay and lipid analysis were performed as described in Materials and Methods. Abbreviations: 1-OH-TAG, monoricinoleoyl-TAG; 1-OH-1-ep-TAG, monoricinoleoyl-monovernoloyl-TAG; OH-FA, unesterified ricinoleic acid.
Figure 3
Lipid content (A and B) and PDAT activity (C) in PDAT-overexpressing yeast cells. The PDAT gene in the plasmid pUS4 was overexpressed from the galactose-induced GAL1 promoter in the wild-type strain W303-1A (10). Its expression was induced after 2 h (A) or 25 h (B) of growth as described in Materials and Methods. The amount of TAG (open bar), polar lipids (hatched bar), sterol esters (filled bar), and other lipids (striped bar) of these cells is presented as μmol of fatty acids per mg of dry weight. The data shown are the mean values of results with three independent yeast cultures. (C) In vitro synthesis of TAG by microsomes prepared from yeast cells, cultivated as in A, containing either the empty vector (vector) or the PDAT plasmid (+PDAT). The substrate lipids dioleoyl-DAG (2.5 nmol) and _sn_-1-oleoyl-_sn_-2-[14C]oleoyl-PC (2 nmol) were added to aliquots of microsomes, which were then incubated for 10 min at 30°C. The results shown are the mean values of two experiments.
Figure 4
Substrate specificity of yeast PDAT. (A) _sn_-position specificity of yeast PDAT regarding the acyl donor substrate. Dioleoyl-DAG (2.5 nmol) together with 4 nmol of _sn_-1-[14C]oleoyl-_sn_-2-[14C]oleoyl-PC (di-[14C]-PC), _sn_-1-[14C]oleoyl-_sn_-2-oleoyl-PC (_sn_1-[14C]-PC), or _sn_-1-oleoyl-_sn_-2-[14C]oleoyl-PC (_sn_2-[14C]-PC) was used as substrate. (B) Specificity of yeast PDAT regarding the phospholipid headgroup and the acyl composition of the phospholipid as well as the DAG. Dioleoyl-DAG (2.5 nmol) together with 4 nmol of _sn_-1-oleoyl-_sn_-2-[14C]oleoyl-PC (oleoyl-PC), _sn_-1-oleoyl-_sn_-2-[14C]oleoyl-PE (oleoyl-PE), _sn_-1-oleoyl-_sn_-2-[14C]ricinoleoyl-PC (ricinoleoyl-PC), or _sn_-1-oleoyl-_sn_-2-[14C]vernoloyl-PC (vernoloyl-PC) was used as substrate. In the experiments presented in the two bars to the far right, 2.5 nmol of monoricinoleoyl-DAG (ricinoleoyl-DAG) or monovernoloyl-DAG (vernoloyl-DAG) were used together with 4 nmol of _sn_-1-oleoyl-sn_-2-[14C]oleoyl-PC. Microsomes from W303-1A cells overexpressing the PDAT gene, as described in Fig. 3_A, were incubated at 30°C for 10 min (A) or 90 min (B). The synthesis of radiolabeled TAG (solid bars) and lyso-PC (LPC, open bars) are the mean values of two experiments.
Figure 5
Evolutionary dendrogram showing LCAT- and PDAT-related proteins from different eukaryotes. The dendrogram was calculated from aligned protein sequences corresponding to amino acid residues 174–335 in yeast PDAT. The
clustalx
multiple alignment program (28) was used with default settings to align the sequences and compute pairwise alignment scores. An unrooted tree was then obtained from these scores by using the neighbor-joining method (29), with correction for multiple substitutions and exclusion of gapped positions.
Similar articles
- The Phospholipid:Diacylglycerol Acyltransferase-Mediated Acyl-Coenzyme A-Independent Pathway Efficiently Diverts Fatty Acid Flux from Phospholipid into Triacylglycerol in Escherichia coli.
Wang L, Jiang S, Chen WC, Zhou XR, Huang TX, Huang FH, Wan X. Wang L, et al. Appl Environ Microbiol. 2020 Sep 1;86(18):e00999-20. doi: 10.1128/AEM.00999-20. Print 2020 Sep 1. Appl Environ Microbiol. 2020. PMID: 32680871 Free PMC article. - Activities of acyl-CoA:diacylglycerol acyltransferase (DGAT) and phospholipid:diacylglycerol acyltransferase (PDAT) in microsomal preparations of developing sunflower and safflower seeds.
Banaś W, Sanchez Garcia A, Banaś A, Stymne S. Banaś W, et al. Planta. 2013 Jun;237(6):1627-36. doi: 10.1007/s00425-013-1870-8. Epub 2013 Mar 29. Planta. 2013. PMID: 23539042 Free PMC article. - Cloning and functional characterization of a phospholipid:diacylglycerol acyltransferase from Arabidopsis.
Ståhl U, Carlsson AS, Lenman M, Dahlqvist A, Huang B, Banas W, Banas A, Stymne S. Ståhl U, et al. Plant Physiol. 2004 Jul;135(3):1324-35. doi: 10.1104/pp.104.044354. Epub 2004 Jul 9. Plant Physiol. 2004. PMID: 15247387 Free PMC article. - Properties and Biotechnological Applications of Acyl-CoA:diacylglycerol Acyltransferase and Phospholipid:diacylglycerol Acyltransferase from Terrestrial Plants and Microalgae.
Xu Y, Caldo KMP, Pal-Nath D, Ozga J, Lemieux MJ, Weselake RJ, Chen G. Xu Y, et al. Lipids. 2018 Jul;53(7):663-688. doi: 10.1002/lipd.12081. Epub 2018 Sep 25. Lipids. 2018. PMID: 30252128 Review. - Physiological Functions of Phospholipid:Diacylglycerol Acyltransferases.
Sah SK, Fan J, Blanford J, Shanklin J, Xu C. Sah SK, et al. Plant Cell Physiol. 2024 Jun 27;65(6):863-871. doi: 10.1093/pcp/pcad106. Plant Cell Physiol. 2024. PMID: 37702708 Review.
Cited by
- A mechanism implicating plastoglobules in thylakoid disassembly during senescence and nitrogen starvation.
Besagni C, Kessler F. Besagni C, et al. Planta. 2013 Feb;237(2):463-70. doi: 10.1007/s00425-012-1813-9. Epub 2012 Nov 28. Planta. 2013. PMID: 23187680 Review. - Acyl-lipid metabolism.
Li-Beisson Y, Shorrosh B, Beisson F, Andersson MX, Arondel V, Bates PD, Baud S, Bird D, Debono A, Durrett TP, Franke RB, Graham IA, Katayama K, Kelly AA, Larson T, Markham JE, Miquel M, Molina I, Nishida I, Rowland O, Samuels L, Schmid KM, Wada H, Welti R, Xu C, Zallot R, Ohlrogge J. Li-Beisson Y, et al. Arabidopsis Book. 2010;8:e0133. doi: 10.1199/tab.0133. Epub 2010 Jun 11. Arabidopsis Book. 2010. PMID: 22303259 Free PMC article. - Large fluxes of fatty acids from membranes to triacylglycerol and back during N-deprivation and recovery in Chlamydomonas.
Young DY, Shachar-Hill Y. Young DY, et al. Plant Physiol. 2021 Apr 2;185(3):796-814. doi: 10.1093/plphys/kiaa071. Plant Physiol. 2021. PMID: 33822218 Free PMC article. - The Phospholipid:Diacylglycerol Acyltransferase-Mediated Acyl-Coenzyme A-Independent Pathway Efficiently Diverts Fatty Acid Flux from Phospholipid into Triacylglycerol in Escherichia coli.
Wang L, Jiang S, Chen WC, Zhou XR, Huang TX, Huang FH, Wan X. Wang L, et al. Appl Environ Microbiol. 2020 Sep 1;86(18):e00999-20. doi: 10.1128/AEM.00999-20. Print 2020 Sep 1. Appl Environ Microbiol. 2020. PMID: 32680871 Free PMC article. - Lipid droplet formation on opposing sides of the endoplasmic reticulum.
Sturley SL, Hussain MM. Sturley SL, et al. J Lipid Res. 2012 Sep;53(9):1800-10. doi: 10.1194/jlr.R028290. Epub 2012 Jun 14. J Lipid Res. 2012. PMID: 22701043 Free PMC article. Review.
References
- Bell R M, Coleman R A. Annu Rev Biochem. 1980;49:459–487. - PubMed
- Stymne S, Stobart K. In: The Biochemistry of Plants: A Comprehensive Treatise. Stumpf P K, editor. Vol. 9. New York: Academic; 1987. pp. 175–214.
- Hobbs D H, Lu C, Hills M J. FEBS Lett. 1999;452:145–149. - PubMed
- Routaboul J M, Benning C, Bechtold N, Caboche M, Lepiniec L. Plant Physiol Biochem. 1999;37:831–840. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous