Yeast nucleoporins involved in passive nuclear envelope permeability - PubMed (original) (raw)
Yeast nucleoporins involved in passive nuclear envelope permeability
N Shulga et al. J Cell Biol. 2000.
Abstract
The vertebrate nuclear pore complex (NPC) harbors an approximately 10-nm diameter diffusion channel that is large enough to admit 50-kD polypeptides. We have analyzed the permeability properties of the Saccharomyces cerevisiae nuclear envelope (NE) using import (NLS) and export (NES) signal-containing green fluorescent protein (GFP) reporters. Compared with wild-type, passive export rates of a classical karyopherin/importin (Kap) Kap60p/Kap95p-targeted NLS-GFP reporter (cNLS-GFP) were significantly faster in nup188-Delta and nup170-Delta cells. Similar results were obtained using two other NLS-GFP reporters, containing either the Kap104p-targeted Nab2p NLS (rgNLS) or the Kap121p-targeted Pho4p NLS (pNLS). Elevated levels of Hsp70 stimulated cNLS-GFP import, but had no effect on the import of rgNLS-GFP. Thus, the role of Hsp70 in NLS-directed import may be NLS- or targeting pathway-specific. Equilibrium sieving limits for the diffusion channel were assessed in vivo using NES-GFP reporters of 36-126 kD and were found to be greater than wild-type in nup188-Delta and nup170-Delta cells. We propose that Nup170p and Nup188p are involved in establishing the functional resting diameter of the NPC's central transport channel.
Figures
Figure 1
Export kinetics of cNLS-GFP as a function of temperature in azide/deoxyglucose. Cells expressing cNLS-GFP were grown at 24°C, pelleted, washed, and suspended in glucose-free SC medium containing 10 mM azide and 10 mM 2-deoxyglucose that had been warmed or chilled at the assay temperature. Export was quantified as described in Materials and Methods.
Figure 2
Steady-state localization of cNLS-GFP in wt, _nup170-_Δ, and _nup188-_Δ cells was determined before induction of GAL1-SSA1 (23°C), after induction (23°C +SSA1), and incubated on ice after induction (0°C +SSA1). GFP fluorescence and Hoechst stain images were obtained by confocal microscopy (see Materials and Methods).
Figure 3
Temperature dependence of cNLS-GFP nuclear accumulation in wt and mutant cells. A, Dynamics of cNLS-GFP localization in wt and _nup188-_Δ cells ± GAL1-SSA1 induction were quantified as described in Materials and Methods. At time = 0, cells were shifted from 23°C to an ice bath (0°C). Wt cells were maintained at 0°C for the duration of the time course. Arrows on the time line indicate when _nup188-_Δ cells were shifted from 0 to 23°C (15 min) and then back to 0°C (30 min). B, Relaxation kinetics of cNLS-GFP redistribution in _nup188-_Δ + SSA1 cells after shifting initial incubation temperature from 30 to 20, 10, or 0°C. C, Export kinetics of cNLS-GFP at 0°C in additional null strains.
Figure 3
Temperature dependence of cNLS-GFP nuclear accumulation in wt and mutant cells. A, Dynamics of cNLS-GFP localization in wt and _nup188-_Δ cells ± GAL1-SSA1 induction were quantified as described in Materials and Methods. At time = 0, cells were shifted from 23°C to an ice bath (0°C). Wt cells were maintained at 0°C for the duration of the time course. Arrows on the time line indicate when _nup188-_Δ cells were shifted from 0 to 23°C (15 min) and then back to 0°C (30 min). B, Relaxation kinetics of cNLS-GFP redistribution in _nup188-_Δ + SSA1 cells after shifting initial incubation temperature from 30 to 20, 10, or 0°C. C, Export kinetics of cNLS-GFP at 0°C in additional null strains.
Figure 4
Import kinetics of cNLS-GFP at 0°C. Wt, _nup170-_Δ, and _nup188-_Δ cells were treated for 40 min with 10 mM 2-deoxyglucose at 30°C to equilibrate cNLS-GFP. Cells were then washed and resuspended in ice-cold complete medium and incubated at 0°C. Cells were harvested at various times and assayed for cNLS-GFP nuclear accumulation as described in Materials and Methods. Also shown are fluorescence (GFP) and light (DIC) images of wt cells before and after 2-deoxyglucose-induced equilibration, and after 20 h on ice.
Figure 4
Import kinetics of cNLS-GFP at 0°C. Wt, _nup170-_Δ, and _nup188-_Δ cells were treated for 40 min with 10 mM 2-deoxyglucose at 30°C to equilibrate cNLS-GFP. Cells were then washed and resuspended in ice-cold complete medium and incubated at 0°C. Cells were harvested at various times and assayed for cNLS-GFP nuclear accumulation as described in Materials and Methods. Also shown are fluorescence (GFP) and light (DIC) images of wt cells before and after 2-deoxyglucose-induced equilibration, and after 20 h on ice.
Figure 5
Effects of chilling, _GAL1_-SSA1 expression, and azide/deoxyglucose on the nuclear localization of rgNLS-GFP in wt, _nup188-_Δ, and _nup170-_Δ cells. A, Localization of rgNLS-GFP in wt, _nup170-_Δ, and _nup188-_Δ cells was determined before induction of GAL1-SSA1 (23°C), after induction (23°C +SSA1), after incubation on ice after induction (0°C +SSA1), and after incubation in azide/deoxyglucose. GFP and DIC images were captured by confocal microscopy (see Materials and Methods). B, Kinetic import assay of rgNLS-GFP in wt and mutant cells after equilibration in azide/deoxyglucose. Import was assayed in _nup170-_Δ and _nup188-_Δ cells ± GAL1-SSA1 induction. Kinetic assays were performed at 37°C as described in Materials and Methods.
Figure 5
Effects of chilling, _GAL1_-SSA1 expression, and azide/deoxyglucose on the nuclear localization of rgNLS-GFP in wt, _nup188-_Δ, and _nup170-_Δ cells. A, Localization of rgNLS-GFP in wt, _nup170-_Δ, and _nup188-_Δ cells was determined before induction of GAL1-SSA1 (23°C), after induction (23°C +SSA1), after incubation on ice after induction (0°C +SSA1), and after incubation in azide/deoxyglucose. GFP and DIC images were captured by confocal microscopy (see Materials and Methods). B, Kinetic import assay of rgNLS-GFP in wt and mutant cells after equilibration in azide/deoxyglucose. Import was assayed in _nup170-_Δ and _nup188-_Δ cells ± GAL1-SSA1 induction. Kinetic assays were performed at 37°C as described in Materials and Methods.
Figure 6
Effects of chilling on the nuclear localization of pNLS-GFP in wt, _nup188-_Δ, and _nup170-_Δ cells. Cells were grown at 23°C and incubated at 0°C as described in Materials and Methods. GFP fluorescence and Hoechst stain images were obtained by confocal microscopy.
Figure 7
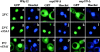
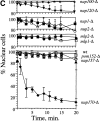
NE sieving limits in wt, _nup188-_Δ, and _nup170-_Δ cells with NES-GFP molecular size probes. A, Localization of NES-GFP66 at 23 and 0°C in wt, _nup188-_Δ, and _nup170-_Δ cells. NES-GFP66 fluorescence (GFP) and Hoechst stain images were obtained by confocal microscopy. B, Time course of reexport of equilibrated NES-GFP66 in _nup188-_Δ cells. _nup188-_Δ cells incubated at 0°C for 1 h were transferred to a slide and the localization of NES-GFP66 observed as the cells warmed to room temperature (∼23°C). C, Quantification of nucleocytoplasmic distributions of NES-GFP size probes in wt, _nup188-_Δ, and _nup170-_Δ cells at 23 and 0°C. NES-GFP66, 81-, and 126-kD probes were expressed in wt and mutant cells at 23°C and then shifted to 0°C for 1 h. The top value in each ratio is the [C]/[N] ratio at 23°C and the bottom value is the [C]/[N] ratio at 0°C (the values of the ratios of these ratios are indicated to the right). In each case, nuclear and cytoplasmic levels of GFP fluorescence ([C]/[N]) at 23 and 0°C were quantified using the averaged pixel density of three different areas within the nuclei and cytoplasms of 10–15 different cells using the region tool in MetaMorph.
Similar articles
- Interactions between a nuclear transporter and a subset of nuclear pore complex proteins depend on Ran GTPase.
Seedorf M, Damelin M, Kahana J, Taura T, Silver PA. Seedorf M, et al. Mol Cell Biol. 1999 Feb;19(2):1547-57. doi: 10.1128/MCB.19.2.1547. Mol Cell Biol. 1999. PMID: 9891088 Free PMC article. - Specific binding of the karyopherin Kap121p to a subunit of the nuclear pore complex containing Nup53p, Nup59p, and Nup170p.
Marelli M, Aitchison JD, Wozniak RW. Marelli M, et al. J Cell Biol. 1998 Dec 28;143(7):1813-30. doi: 10.1083/jcb.143.7.1813. J Cell Biol. 1998. PMID: 9864357 Free PMC article. - An essential nuclear envelope integral membrane protein, Brr6p, required for nuclear transport.
de Bruyn Kops A, Guthrie C. de Bruyn Kops A, et al. EMBO J. 2001 Aug 1;20(15):4183-93. doi: 10.1093/emboj/20.15.4183. EMBO J. 2001. PMID: 11483521 Free PMC article. - In vivo nuclear transport kinetics in Saccharomyces cerevisiae.
Roberts PM, Goldfarb DS. Roberts PM, et al. Methods Cell Biol. 1998;53:545-57. doi: 10.1016/s0091-679x(08)60894-8. Methods Cell Biol. 1998. PMID: 9348524 Review. - The nuclear pore complex.
Hurt EC. Hurt EC. FEBS Lett. 1993 Jun 28;325(1-2):76-80. doi: 10.1016/0014-5793(93)81417-x. FEBS Lett. 1993. PMID: 8513897 Review.
Cited by
- The nucleoporin Nup188 controls passage of membrane proteins across the nuclear pore complex.
Theerthagiri G, Eisenhardt N, Schwarz H, Antonin W. Theerthagiri G, et al. J Cell Biol. 2010 Jun 28;189(7):1129-42. doi: 10.1083/jcb.200912045. Epub 2010 Jun 21. J Cell Biol. 2010. PMID: 20566687 Free PMC article. - Global motions of the nuclear pore complex: insights from elastic network models.
Lezon TR, Sali A, Bahar I. Lezon TR, et al. PLoS Comput Biol. 2009 Sep;5(9):e1000496. doi: 10.1371/journal.pcbi.1000496. Epub 2009 Sep 4. PLoS Comput Biol. 2009. PMID: 19730674 Free PMC article. - Caenorhabditis elegans nucleoporins Nup93 and Nup205 determine the limit of nuclear pore complex size exclusion in vivo.
Galy V, Mattaj IW, Askjaer P. Galy V, et al. Mol Biol Cell. 2003 Dec;14(12):5104-15. doi: 10.1091/mbc.e03-04-0237. Epub 2003 Aug 22. Mol Biol Cell. 2003. PMID: 12937276 Free PMC article. - Size-dependent leak of soluble and membrane proteins through the yeast nuclear pore complex.
Popken P, Ghavami A, Onck PR, Poolman B, Veenhoff LM. Popken P, et al. Mol Biol Cell. 2015 Apr 1;26(7):1386-94. doi: 10.1091/mbc.E14-07-1175. Epub 2015 Jan 28. Mol Biol Cell. 2015. PMID: 25631821 Free PMC article. - Binding dynamics of structural nucleoporins govern nuclear pore complex permeability and may mediate channel gating.
Shulga N, Goldfarb DS. Shulga N, et al. Mol Cell Biol. 2003 Jan;23(2):534-42. doi: 10.1128/MCB.23.2.534-542.2003. Mol Cell Biol. 2003. PMID: 12509452 Free PMC article.
References
- Adam S.A. Transport pathways of macromolecules between the nucleus and the cytoplasm. Curr. Opin. Cell Biol. 1999;11:402–406. - PubMed
- Aitchison J.D., Rout M.P., Marelli M., Blobel G., Wozniak R.W. Two novel related yeast nucleoporins Nup170p and Nup157pcomplementation with the vertebrate homologue Nup155p and functional interactions with the yeast nuclear pore membrane protein Pom152p. J. Cell Biol. 1995;131:1133–1148. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases