Translational control of viral gene expression in eukaryotes - PubMed (original) (raw)
Review
Translational control of viral gene expression in eukaryotes
M Gale Jr et al. Microbiol Mol Biol Rev. 2000 Jun.
Abstract
As obligate intracellular parasites, viruses rely exclusively on the translational machinery of the host cell for the synthesis of viral proteins. This relationship has imposed numerous challenges on both the infecting virus and the host cell. Importantly, viruses must compete with the endogenous transcripts of the host cell for the translation of viral mRNA. Eukaryotic viruses have thus evolved diverse mechanisms to ensure translational efficiency of viral mRNA above and beyond that of cellular mRNA. Mechanisms that facilitate the efficient and selective translation of viral mRNA may be inherent in the structure of the viral nucleic acid itself and can involve the recruitment and/or modification of specific host factors. These processes serve to redirect the translation apparatus to favor viral transcripts, and they often come at the expense of the host cell. Accordingly, eukaryotic cells have developed antiviral countermeasures to target the translational machinery and disrupt protein synthesis during the course of virus infection. Not to be outdone, many viruses have answered these countermeasures with their own mechanisms to disrupt cellular antiviral pathways, thereby ensuring the uncompromised translation of virion proteins. Here we review the varied and complex translational programs employed by eukaryotic viruses. We discuss how these translational strategies have been incorporated into the virus life cycle and examine how such programming contributes to the pathogenesis of the host cell.
Figures
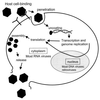
FIG. 1
General model of eukaryotic viral replication. Viral particles are shown in black. Viruses recognize their target cell through interaction with specific receptors and/or other components on the cell membrane. Interaction with the host cell induces cell membrane penetration and virion internalization. Virion uncoating releases the viral genome, whereupon it is available for transcription and translation. Poxviruses and the RNA viruses (with the exception of retroviruses) replicate in the cytoplasm. Transcription of all other DNA viruses takes place in the nucleus. Transcription and genome replication are followed by the cytoplasmic stages of mRNA translation and virion assembly. Release of mature virions may include membrane lysis and death of the host cell. Adapted with modification from reference .
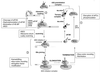
FIG. 2
Schematic illustration of eukaryotic mRNA translation and major sites of viral regulation. Details of the translation process are described in the text. Translation initiation factors are shown by their letter and number designation (335). 40S and 80S denote the small ribosomal subunit and the elongating ribosome, respectively. 1, Ternary-complex formation and assembly of the 43S pre-initiation complex; 2, assembly of the cap-binding complex and ribosome loading onto the mRNA; 3, Ribosome scanning to the first AUG codon, recycling of eIF2-GDP, and joining of the 60S ribosomal subunit. TER denotes a translation termination codon. Major sites for viral control of translation and mechanisms of translation control are shown in the surrounding boxes. Not shown is the mRNA 3′ UTR, which can also influence translational efficiency. aa, amino acid.
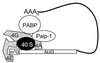
FIG. 3
Model of the closed-loop mRNA translation complex. The mRNA-bound eIF4F initiation complex interacts with the 3′ end of the mRNA via PABP. Poly(A) sequences within the 3′ UTR direct PABP binding to the mRNA. PABP mediates interaction with the cap-binding complex either directly through eIF4G (4G) (452) or indirectly through an eIF4G, eIF4A (4A)-dependent interaction with Paip-1 (75). Assembly of the closed-loop complex may stabilize the interaction of the 40S ribosomal subunit with the mRNA (225, 401).
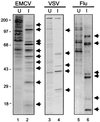
FIG. 4
Virus-induced shutoff of host cell protein synthesis. Murine NIH 3T3 cells (lanes 1 to 4) or Madin-Darby bovine kidney cells (lanes 5 and 6) were mock infected (U) or infected (I), respectively, with EMCV (lanes 1 and 2), VSV (lanes 3 and 4), or influenza virus (lanes 5 and 6). To visualize the virus-induced host shutoff of protein synthesis and the concomitant shift to viral protein synthesis, proteins were biosynthetically labeled by the addition of [35S]methionine to the culture medium. Protein equivalents from mock-infected and virus-infected cells were separated by gel electrophoresis and visualized by autoradiography of the dried gel. Arrows denote the positions of viral proteins. The positions of molecular mass standards are indicated in kilodaltons.
FIG. 5
Viral mechanisms of translational programming. The top diagram shows structural features of a representative mRNA containing a m7G cap and consisting of a series of overlapping and nonoverlapping reading frames (denoted by rectangles). The first reading frame is indicated by an AUG initiation codon and is preceded by a 5′ UTR. Upright arrows indicate translation initiation of the corresponding reading frame(s), resulting from the mechanisms listed at left. Specific viruses examples presented within the text are listed at right. Depiction of the IRES and ribosome shunt includes the relevant stem-loop structures within the 5′ UTR. Arrow shows ribosome bypass, or shunting, around the stem-loop. Figure adapted with modification from reference .
FIG. 6
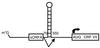
IRES structure. Structural representation of the 5′ UTR from EMCV (top left), poliovirus (top right), HAV (bottom left), and HCV (bottom right). The major stem-loops are labeled according to previous designations (42, 193, 206, 483). The region encompassing the IRES is underlined. Pyrimidine-rich sequence elements are shown as solid rectangles. AUG denotes the position of the translation initiation codon. The box on the HCV IRES denotes the core protein-coding region.
FIG. 7
The CaMV ribosome shunt. Specific details are described in the text. The relative positions of sORFA, the major stem-loop, and the gene VII ORF of the 35S RNA are shown. Ribosome shunting (denoted by the lower arrow) occurs around the major stem-loop encompassing nt 70 to 550 (196). After terminating sORFA translation, the 40S ribosomal subunit is shunted around the major stem-loop structure and resumes scanning to initiate translation at the ORF VII AUG codon (upper arrow).
FIG. 8
Leaky scanning and translational frameshifting during retroviral mRNA translation. (A) Leaky scanning during HIV mRNA translation accounts for synthesis of the Vpu and Env proteins. The HIV gene structure (upper) consists of several overlapping cistrons encoded within a heterogeneous array of mRNAs (438). Synthesis of the Vpu and Env proteins (lower diagram) of HIV proceeds via a leaky-scanning mechanism in which the scanning ribosome bypasses the weak vpu AUG codon (shown) to initiate translation from the env AUG codon (bent arrow). Translation initiation occurs at the vpu AUG codon at a low frequency and may account for the stoichiometric ratios of Vpu and Env protein accumulation during HIV infection (413). (B) Translational frameshifting at the gag-pro junction during MMTV mRNA translation. An RNA pseudoknot near the 3′ end of the gag gene causes the elongating ribosome to pause at the gag-pro slippery sequence element. As a result, the mRNA slips backward by 1 nt and the ribosome-bound tRNAs mediate new anticodon base pairing in the −1 reading frame (12). Frameshifting is favored by weak codon-tRNA anticodon base pairing in the original reading frame and strong base pairing in the new reading frame. Synthesis of the entire Gag-Pro-Pol polyprotein is facilitated by a second frameshift at the downstream pro-pol slippery site. Model adapted from reference .
FIG. 9
CMV control of gp48 synthesis by inefficient termination of uORF2. During CMV infection the viral gp48 glycoprotein is translated from the third of three cistrons (rectangles) within the gpUL4 mRNA. 1, gp48 synthesis is facilitated by a leaky-scanning mechanism in which the scanning ribosome bypasses the weak upstream AUG codons within the gpUL4 mRNA to initiate translation at the gp48 AUG (indicated by arrow); 2, translation initiation at the uORF2 AUG occurs at a low frequency and results in control of gp48 translation. Synthesis of the uORF2 peptide produces a stable peptide-tRNApro-ribosome complex that prevents peptide release and stalls the elongating ribosome at the uORF2 termination codon. As a result, ribosome scanning and reinitiation at the downstream gp48 AUG codon is blocked.
FIG. 10
Functional recoding at the MuLV gag-pol junction. Synthesis of the MuLV Gag protein terminates at the gag stop codon (top). However, approximately 5% of the time, the elongating ribosome will read through the gag stop codon to produce the Gag-Pol polyprotein (bottom). During this process, the gag stop codon is redefined to encode glutamine and is shown by the presence of tRNAGln at the redefined UAG codon. Stop codon redefinition is dependent on specific downstream sequences and a 3′-proximal pseudoknot structure. The elongating ribosome eventually melts out the pseudoknot to complete Gag-Pol synthesis. Low-frequency gag-pol stop codon redefinition is essential for MuLV replication. Figure adapted with modification from references and .
FIG. 11
Translational control during HSV-1 infection. 1, Infection with HSV-1 induces a rapid shutoff of host cell protein synthesis, due in part to the actions of the viral VHS and ICP27 proteins, which indirectly affect mRNA translation. VHS-induced translational shutoff may be directly modulated through the viral VP16 protein. 2, Repression of VHS by VP16 may contribute to the selective translation of viral mRNAs. The viral U(L)13 and ICP0 proteins may similarly modulate viral mRNA translation by phosphorylating and binding, respectively, to host EF1-δ. During infection, HSV-1 ensures that the host cell remains translationally competent by blocking the phosphorylation of eIF2α. Disruption of eIF2α phosphorylation may occur through the actions of the viral Us11 protein, which prevents PKR activation, and by the viral γ134.5 gene product, which directs the dephosphorylation of eIF2α by PPIα. Finally, viral modulation of ribosomal protein S6 phosphorylation may contribute to the sustained translation of host 5′ TOP mRNAs, including those that encode ribosomal proteins. Sustained ribosomal protein synthesis and disruption of eIF2α phosphorylation facilitates HSV persistence by supporting viral mRNA translation.
FIG. 12
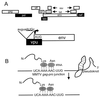
GRSF-1 binds the 5′ UTR of influenza virus mRNAs to stimulate viral mRNA translation. (A) Specific interaction of GRSF-1 with the 5′ UTR of influenza virus NP mRNA. The 5′ UTR of the viral NP mRNA has been functionally divided into at least three contiguous sequence elements (denoted by A to C) (361). The 12-nt A region (underlined) is conserved in all influenza virus mRNAs (274). Dashed lines indicate deleted regions within the respective 5′ UTR constructs used to assess GRSF-1 binding. AINV refers to the NP 5′ UTR in which the sequence of the A region has been inverted. The sequence of a control 5′ UTR from the cellular SEAP gene is shown at the bottom. The A and B regions of the NP 5′ UTR are both required for binding to GRSF-1 (362). (B) Competition for GRSF-1 selectively blocks translation from the NP 5′ UTR in a cell-free system. To determine the contribution of GRSF-1 to influenza virus mRNA translation, the expression of a luciferase reporter protein was placed under control of the NP 5′ UTR. Relative luciferase activity from the resulting NP-Luc construct was assessed in a cell-free system in the absence (No comp) or presence of competitor oligonucleotides encoding the NP 5′ UTR (NP), NP-A, SEAP, or NP-C. (C) Structural representation of GRSF-1. The three RRMs are indicated in black. The 41-amino-acid acidic domain is shown in gray.
FIG. 13
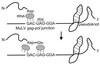
Models for the selective translation of influenza virus mRNAs by GRSF-1. GRSF-1 binding may be facilitated by the AGGGU sequence spanning the junction of the A and B regions within the NP 5′ UTR (362). The two models shown reflect the cap dependency and requirement for eIF4G in influenza virus mRNA translation. (I) GRSF-1 may participate in recruitment of the cap-binding complex to the mRNA by physically interacting with one or more components of eIF4F. Active recruitment of the eIF4F complex by GRSF-1 may overcome the reduced affinity of phosphorylated eIF4E for the m7G cap that occurs during influenza virus infection (110). As a result, GRSF-1 may enhance ribosome binding to the mRNA by increasing the stability of the eIF4F-mRNA complex (bottom). (II) Alternatively, GRSF-1 may allow viral mRNA translation to proceed independently of eIF4E by directly participating in ribosome recruitment with the other components of the eIF4F complex. In this model, GRSF-1 would function in the absence of eIF4E to promote ribosome binding to the mRNA (bottom), thereby avoiding the limitations on translation due to eIF4E phosphorylation. 4E, eIF4E; 4G, eIF4G, 4A, eIF4A.
FIG. 14
Domain structure of eIF4G. The arrow points to the site of cleavage by poliovirus 2A-Pro. Shaded areas indicate the eIF4E-, eIF3-, and eIF4A-binding domains. The bar indicates the region responsible for binding to Mnk1. Numbering refers to the prototypic eIF4GI (153).
FIG. 15
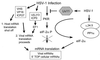
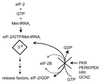
Translational control by eIF2α phosphorylation. eIF2 is composed of several subunits, including the alpha subunit (eIF2α), which is targeted for phosphorylation by the eIF2α protein kinases. eIF2 participates in mRNA translation by delivering the Met-tRNAi to the incoming 40S ribosomal subunit in the form of an eIF2–GTP–Met-tRNAi ternary complex (3°) (335). After Met-tRNAi binding, eIF2 is released from the complex in an inactive state bound to GDP. The bound GDP is recycled for GTP by the eIF2B guanine nucleotide exchange factor (dotted and gray lines), and the initiation process continues. PKR, PERK, PEK, HRI, and GCN2 can block the guanine nucleotide exchange reaction by phosphorylating serine 51 of eIF2α. As a result, the pool of functional eIF2 is depleted and translation arrests at the initiation stage.
FIG. 16
Structure and activation of PKR. (A) Domain structure of PKR and sites of viral regulation. The PKR regulatory domain spans amino acids 1 to 264 and includes two dsRBMs (indicated in black) which mediate binding to activator dsRNA. The protein kinase catalytic domain comprises the PKR C terminus (amino acids 265 to 551) and contains the 11 subdomains (denoted by Roman numerals) conserved in all eukaryotic protein kinases (176). Bars indicate the regions that participate in dsRNA binding, dimerization, and substrate interaction. Virus-directed inhibitors that target PKR function are listed beneath their specific target sites and are referenced in Table 4. (B) PKR activation. PKR is transcriptionally induced by IFNs and becomes active through a process of dsRNA binding and dimerization. Once active, PKR can phosphorylate eIF2α. Within a virus-infected cell, PKR-mediated phosphorylation of eIF2α results in a block in protein synthesis, cell growth arrest, and inhibition of viral replication (241, 242). Additionally, PKR may participate in the regulation of other IFN-induced genes by signaling the phosphorylation of IkB or the activation of IRF-1 (277).
FIG. 17
Translational control by HCV. (A) Structural representation of the HCV polyprotein. The individual positions of the polyprotein cleavage products are shown. NS5A (black region) from IFN-resistant HCV can bind and repress PKR (129, 133). (B) NS5A blocks PKR-dependent eIF2α phosphorylation. eIF2α phosphorylation from control (Neo) NIH 3T3 cell lines and those stably expressing NS5A from IFN-resistant HCV (NS5A-1A) or a nonfunctional NS5A mutant (ΔISDR) was assessed by single-dimension isoelectric focusing of cell extracts and anti-eIF2α immunoblot analysis (133). Cells were mock infected (lanes 1, 3, and 5) or infected with VSV (lanes 2, 4, and 6). Arrows denote the positions hypo- and hyper-phosphorylated isoforms of eIF2α (eIF2α and eIF2αP, respectively). Hyperphosphorylated eIF2α is phosphorylated on serine 51 by PKR and is sufficient to block mRNA translation (61, 89).
Similar articles
- Translation initiation and viral tricks.
Schneider RJ, Mohr I. Schneider RJ, et al. Trends Biochem Sci. 2003 Mar;28(3):130-6. doi: 10.1016/S0968-0004(03)00029-X. Trends Biochem Sci. 2003. PMID: 12633992 Review. - Non-Canonical Translation Initiation Mechanisms Employed by Eukaryotic Viral mRNAs.
Sorokin II, Vassilenko KS, Terenin IM, Kalinina NO, Agol VI, Dmitriev SE. Sorokin II, et al. Biochemistry (Mosc). 2021 Sep;86(9):1060-1094. doi: 10.1134/S0006297921090042. Biochemistry (Mosc). 2021. PMID: 34565312 Free PMC article. Review. - Tinkering with translation: protein synthesis in virus-infected cells.
Walsh D, Mathews MB, Mohr I. Walsh D, et al. Cold Spring Harb Perspect Biol. 2013 Jan 1;5(1):a012351. doi: 10.1101/cshperspect.a012351. Cold Spring Harb Perspect Biol. 2013. PMID: 23209131 Free PMC article. Review. - Regulation of Tacaribe Mammarenavirus Translation: Positive 5' and Negative 3' Elements and Role of Key Cellular Factors.
Foscaldi S, D'Antuono A, Noval MG, de Prat Gay G, Scolaro L, Lopez N. Foscaldi S, et al. J Virol. 2017 Jun 26;91(14):e00084-17. doi: 10.1128/JVI.00084-17. Print 2017 Jul 15. J Virol. 2017. PMID: 28468879 Free PMC article. - So similar, yet so different: selective translation of capped and polyadenylated viral mRNAs in the influenza virus infected cell.
Yángüez E, Nieto A. Yángüez E, et al. Virus Res. 2011 Mar;156(1-2):1-12. doi: 10.1016/j.virusres.2010.12.016. Epub 2010 Dec 31. Virus Res. 2011. PMID: 21195735 Review.
Cited by
- Viral PIC-pocketing: RSV sequestration of translational preinitiation complexes into bi-phasic biomolecular condensates.
Jobe F, Kelly JT, Simpson J, Wells J, Armstrong SD, Spick M, Lacey E, Logan L, Geifman N, Hawes P, Bailey D. Jobe F, et al. J Virol. 2024 Mar 19;98(3):e0015324. doi: 10.1128/jvi.00153-24. Epub 2024 Feb 29. J Virol. 2024. PMID: 38421168 Free PMC article. - Dengue virus utilizes a novel strategy for translation initiation when cap-dependent translation is inhibited.
Edgil D, Polacek C, Harris E. Edgil D, et al. J Virol. 2006 Mar;80(6):2976-86. doi: 10.1128/JVI.80.6.2976-2986.2006. J Virol. 2006. PMID: 16501107 Free PMC article. - Characterization of two distinct RNA domains that regulate translation of the Drosophila gypsy retroelement.
Ronfort C, De Breyne S, Sandrin V, Darlix JL, Ohlmann T. Ronfort C, et al. RNA. 2004 Mar;10(3):504-15. doi: 10.1261/rna.5185604. RNA. 2004. PMID: 14970395 Free PMC article. - Translation of duck hepatitis B virus reverse transcriptase by ribosomal shunting.
Sen N, Cao F, Tavis JE. Sen N, et al. J Virol. 2004 Nov;78(21):11751-7. doi: 10.1128/JVI.78.21.11751-11757.2004. J Virol. 2004. PMID: 15479816 Free PMC article. - Dual functions of Rift Valley fever virus NSs protein: inhibition of host mRNA transcription and post-transcriptional downregulation of protein kinase PKR.
Ikegami T, Narayanan K, Won S, Kamitani W, Peters CJ, Makino S. Ikegami T, et al. Ann N Y Acad Sci. 2009 Sep;1171 Suppl 1(Suppl 1):E75-85. doi: 10.1111/j.1749-6632.2009.05054.x. Ann N Y Acad Sci. 2009. PMID: 19751406 Free PMC article.
References
- Agol V I. Virus robustness and perseverance. Mol Biol. 1998;32:44–49. - PubMed
- Alama A, Barbieri F, Cagnoli M, Schettini G. Antisense oligonucleotides as therapeutic agents. Pharmacol Res. 1997;36:171–178. - PubMed
- Ali N, Wang C, Siddiqui A. Translation of hepatitis C virus genome. Princess Takamatsu Symp. 1995;25:99–110. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P51 RR000166/RR/NCRR NIH HHS/United States
- AI22646/AI/NIAID NIH HHS/United States
- R01 AI022646/AI/NIAID NIH HHS/United States
- RR00166/RR/NCRR NIH HHS/United States
- AI41629/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials