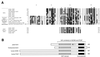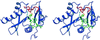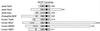Acetylation of histones and transcription-related factors - PubMed (original) (raw)
Review
Acetylation of histones and transcription-related factors
D E Sterner et al. Microbiol Mol Biol Rev. 2000 Jun.
Abstract
The state of chromatin (the packaging of DNA in eukaryotes) has long been recognized to have major effects on levels of gene expression, and numerous chromatin-altering strategies-including ATP-dependent remodeling and histone modification-are employed in the cell to bring about transcriptional regulation. Of these, histone acetylation is one of the best characterized, as recent years have seen the identification and further study of many histone acetyltransferase (HAT) proteins and their associated complexes. Interestingly, most of these proteins were previously shown to have coactivator or other transcription-related functions. Confirmed and putative HAT proteins have been identified from various organisms from yeast to humans, and they include Gcn5-related N-acetyltransferase (GNAT) superfamily members Gcn5, PCAF, Elp3, Hpa2, and Hat1: MYST proteins Sas2, Sas3, Esa1, MOF, Tip60, MOZ, MORF, and HBO1; global coactivators p300 and CREB-binding protein; nuclear receptor coactivators SRC-1, ACTR, and TIF2; TATA-binding protein-associated factor TAF(II)250 and its homologs; and subunits of RNA polymerase III general factor TFIIIC. The acetylation and transcriptional functions of these HATs and the native complexes containing them (such as yeast SAGA, NuA4, and possibly analogous human complexes) are discussed. In addition, some of these HATs are also known to modify certain nonhistone transcription-related proteins, including high-mobility-group chromatin proteins, activators such as p53, coactivators, and general factors. Thus, we also detail these known factor acetyltransferase (FAT) substrates and the demonstrated or potential roles of their acetylation in transcriptional processes.
Figures
FIG. 1
Similarities of GNAT (Gcn5-related _N_-acetyltransferase) superfamily members. (A) Alignment of GNAT homology motifs A, B, C, and D for HATs and representatives of other types of acetyltransferases. Reversed type indicates consensus sequence residues, as determined by Neuwald and Landsman (174), and solid circles mark residues that are particularly conserved throughout the superfamily. The asterisk indicates the glutamate residue known to be critical for HAT catalysis in yeast Gcn5. At the bottom are several members of another acetyltransferase family, the MYST proteins, that share just the A motif. (B) Alignment of proteins from the Gcn5 subgroup of the GNAT superfamily, showing the location of the HAT domain and bromodomain. Shown at the top is the overall homology region shared by all four proteins, with the similarity between the yeast and human proteins indicated; Tetrahymena Gcn5 has 62% similarity with the three others over this same region. The A2 label designates a region in yeast Gcn5 known to interact with the adaptor protein Ada2 (37). In addition, the N-terminal region of PCAF has been generally defined as its site of interaction with p300/CBP and nuclear receptor coactivator SRC-1 (130, 276); this region and the HAT domain also interact with viral protein E1A (40, 193).
FIG. 2
Primary histone acetylation specifities of some of the known HAT proteins in vitro. Shown are the amino acid sequences for the N-terminal tail regions of human histones H2A, H2B, H3, and H4, with lysine residues numbered and arrows indicating the predominant sites used by various HATs in in vitro experiments. TAFII230 is the Drosophila homolog of human TAFII250 used in site specificity determinations. The above H3 and H4 sequences are nearly identical to those of S. cerevisiae and Drosophila. Specific but relatively nonpreferred or minor sites for certain HATs are not indicated, for example, H4 lysine-8 for Gcn5 and PCAF. It should be noted that lysine specificities may be somewhat expanded or restricted with native HAT complexes and/or on nucleosomal substrates.
FIG. 3
Stereo diagram of the structure of a HAT domain bound to its substrates. Shown is the GNAT superfamily protein Tetrahymena Gcn5 (blue) with a histone H3 N-terminal tail peptide (red) and CoA (green) bound to its upper and lower clefts, respectively. At the active site, the glutamate-122 residue (aqua), analogous to yeast Gcn5 glutamate-173, catalyzes the transfer of an acetyl group from acetyl-CoA to the lysine-14 sidechain (orange) of H3 peptide. The N termini of both the Gcn5 protein and the H3 peptide are to the left in diagram, and C-termini are to the right.
FIG. 4
Alignment of the MYST family of HATs and putative HAT proteins. The MYST homology region is indicated, with the acetyl-CoA-binding site, corresponding to GNAT family motif A, shown as a black box. Z, zinc finger motifs: an atypical C2HC motif in the MYST region (Esa1 diverges from this motif), a typical C2HC in the N-terminal region of HBO1, and two adjacent C4HC3 (or PHD) fingers in MOZ and MORF. C, chromo-like domain found in Esa1, MOF, and Tip60.
FIG. 5
Domains and interaction regions of the global coactivator HATs p300/CBP. Labeled below the polypeptide diagram are several domains and sequence motifs, including a bromodomain, the HAT domain, and ZZ and TAZ putative zinc fingers (190). Above are indicated some of the proteins demonstrated to interact with p300/CBP at certain regions. PCAF has been shown to interact with two regions of p300/CBP (130).
FIG. 6
Alignment of the p160 family of mammalian nuclear receptor coactivators. Indicated are the PAS/basic helix-loop-helix homology (bHLH) domain, nuclear receptor interaction regions, and the general area of interaction for coactivators p300/CBP and PCAF. ACTR and SRC-1 each have a HAT domain near their C terminus (42, 220), although the boundaries of these domains have only been approximately defined, and TIF2's domain is inferred by homology.
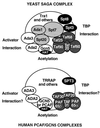
FIG. 7
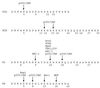
Schematic diagram of known subunits and functions of the yeast SAGA and human PCAF or GCN5 HAT complexes. The yeast SAGA complex (top) contains Gcn5 as its HAT catalytic subunit. SAGA has been shown to interact with acidic activation domains, and this function may be mediated by its adaptor components (Ada2, Ada3, and Gcn5) or possibly through Tra1 or other subunits. Another subset of SAGA proteins (Ada1, Spt7, and Spt20/Ada5) are required for its structural integrity and overall function. The Spt3 and Spt8 subunits have been implicated in interaction with TBP, and the TafII group, which also participate in TFIID, may provide TBP-binding function to SAGA as well. The human SAGA-analogous complexes (bottom) contain either GCN5 or PCAF. The PCAF complex has been more thoroughly studied; it has nucleosome acetylation function, and known subunits include TRRAP, hADA2, hADA3, hSPT3, and TAFIIs/PAFs (PCAF-associated factors)—all homologs of proteins found in SAGA. Activator and TBP interaction functions are hypothesized for these human complexes but have not yet been demonstrated.
Similar articles
- Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex.
Grant PA, Duggan L, Côté J, Roberts SM, Brownell JE, Candau R, Ohba R, Owen-Hughes T, Allis CD, Winston F, Berger SL, Workman JL. Grant PA, et al. Genes Dev. 1997 Jul 1;11(13):1640-50. doi: 10.1101/gad.11.13.1640. Genes Dev. 1997. PMID: 9224714 - Expression, purification, and analysis of MOZ and MORF histone acetyltransferases.
Pelletier N, Champagne N, Lim H, Yang XJ. Pelletier N, et al. Methods. 2003 Sep;31(1):24-32. doi: 10.1016/s1046-2023(03)00084-7. Methods. 2003. PMID: 12893170 - A subset of TAF(II)s are integral components of the SAGA complex required for nucleosome acetylation and transcriptional stimulation.
Grant PA, Schieltz D, Pray-Grant MG, Steger DJ, Reese JC, Yates JR 3rd, Workman JL. Grant PA, et al. Cell. 1998 Jul 10;94(1):45-53. doi: 10.1016/s0092-8674(00)81220-9. Cell. 1998. PMID: 9674426 - The diverse superfamily of lysine acetyltransferases and their roles in leukemia and other diseases.
Yang XJ. Yang XJ. Nucleic Acids Res. 2004 Feb 11;32(3):959-76. doi: 10.1093/nar/gkh252. Print 2004. Nucleic Acids Res. 2004. PMID: 14960713 Free PMC article. Review.
Cited by
- Epigenetic Regulation in Myocardial Fibroblasts and Its Impact on Cardiovascular Diseases.
Komal S, Gao Y, Wang ZM, Yu QW, Wang P, Zhang LR, Han SN. Komal S, et al. Pharmaceuticals (Basel). 2024 Oct 10;17(10):1353. doi: 10.3390/ph17101353. Pharmaceuticals (Basel). 2024. PMID: 39458994 Free PMC article. Review. - Genetic Incorporation of ε-N-2-Hydroxyisobutyryl-lysine into Recombinant Histones.
Xiao H, Xuan W, Shao S, Liu T, Schultz PG. Xiao H, et al. ACS Chem Biol. 2015 Jul 17;10(7):1599-603. doi: 10.1021/cb501055h. Epub 2015 Apr 29. ACS Chem Biol. 2015. PMID: 25909834 Free PMC article. - Role of succinate dehydrogenase deficiency and oncometabolites in gastrointestinal stromal tumors.
Zhao Y, Feng F, Guo QH, Wang YP, Zhao R. Zhao Y, et al. World J Gastroenterol. 2020 Sep 14;26(34):5074-5089. doi: 10.3748/wjg.v26.i34.5074. World J Gastroenterol. 2020. PMID: 32982110 Free PMC article. Review. - Identification of highly selective and potent histone deacetylase 3 inhibitors using click chemistry-based combinatorial fragment assembly.
Suzuki T, Kasuya Y, Itoh Y, Ota Y, Zhan P, Asamitsu K, Nakagawa H, Okamoto T, Miyata N. Suzuki T, et al. PLoS One. 2013 Jul 16;8(7):e68669. doi: 10.1371/journal.pone.0068669. Print 2013. PLoS One. 2013. PMID: 23874714 Free PMC article. - Novel role of PELP1 in regulating chemotherapy response in mutant p53-expressing triple negative breast cancer cells.
Krishnan SR, Nair BC, Sareddy GR, Roy SS, Natarajan M, Suzuki T, Peng Y, Raj G, Vadlamudi RK. Krishnan SR, et al. Breast Cancer Res Treat. 2015 Apr;150(3):487-99. doi: 10.1007/s10549-015-3339-x. Epub 2015 Mar 19. Breast Cancer Res Treat. 2015. PMID: 25788226 Free PMC article.
References
- Adams P D, Kaelin W G J. Transcriptional control by E2F. Semin Cancer Biol. 1995;6:99–108. - PubMed
- Ait-Si-Ali S, Ramirez S, Barre F X, Dkhissi F, Magnaghi-Jaulin L, Girault J A, Robin P, Knibiehler M, Pritchard L L, Ducommun B, Trouche D, Harel-Bellan A. Histone acetyltransferase activity of CBP is controlled by cycle-dependent kinases and oncoprotein E1A. Nature. 1998;396:184–186. - PubMed
- Angus-Hill M L, Dutnall R N, Tafrov S T, Sternglanz R, Ramakrishnan V. Crystal structure of the histone acetyltransferase Hpa2: a tetrameric member of the Gcn5-related N-acetyltransferase superfamily. J Mol Biol. 1999;294:1311–1325. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous