The downstream promoter element DPE appears to be as widely used as the TATA box in Drosophila core promoters - PubMed (original) (raw)
The downstream promoter element DPE appears to be as widely used as the TATA box in Drosophila core promoters
A K Kutach et al. Mol Cell Biol. 2000 Jul.
Abstract
The downstream promoter element (DPE) functions cooperatively with the initiator (Inr) for the binding of TFIID in the transcription of core promoters in the absence of a TATA box. We examined the properties of sequences that can function as a DPE as well as the range of promoters that use the DPE as a core promoter element. By using an in vitro transcription assay, we identified 17 new DPE-dependent promoters and found that all possessed identical spacing between the Inr and DPE. Moreover, mutational analysis indicated that the insertion or deletion of a single nucleotide between the Inr and DPE causes a reduction in transcriptional activity and TFIID binding. To explore the range of sequences that can function as a DPE, we constructed and analyzed randomized promoter libraries. These experiments yielded the DPE functional range set, which represents sequences that contribute to or are compatible with DPE function. We then analyzed the DPE functional range set in conjunction with a Drosophila core promoter database that we compiled from 205 promoters with accurately mapped start sites. Somewhat surprisingly, the DPE sequence motif is as common as the TATA box in Drosophila promoters. There is, in addition, a striking adherence of Inr sequences to the Inr consensus in DPE-containing promoters relative to DPE-less promoters. Furthermore, statistical and biochemical analyses indicated that a G nucleotide between the Inr and DPE contributes to transcription from DPE-containing promoters. Thus, these data reveal that the DPE exhibits a strict spacing requirement yet some sequence flexibility and appears to be as widely used as the TATA box in Drosophila.
Figures
FIG. 1
The distance between the Inr and DPE is strictly maintained in a variety of naturally occurring Drosophila core promoters. (A) In vitro transcription analysis of DPE-containing core promoters. A series of minimal core promoters were constructed with the DNA sequences indicated in the figure. Wild-type (Wt) and DPE mutant (Mut) versions of these promoter constructions were subjected to in vitro transcription and primer extension analysis. The sequences of the mutant promoter constructions as well as the quantitation of the data are given in Table 1. (B) The positioning of DPE-like sequences relative to the Inr is important for DPE function. In Mut1 promoters, DPE-like sequences with improper spacing relative to the Inr are mutated, whereas in Mut2 promoters, DPE sequences with the proper spacing relative to the Inr are mutated. The promoters were subjected to in vitro transcription and primer extension analysis, and the transcriptional activity of each mutant promoter relative to the corresponding wild-type promoter is indicated.
FIG. 2
A single nucleotide alteration in the spacing between the DPE and Inr reduces core promoter activity and binding of purified TFIID. (A) In vitro transcription and primer extension analysis of a series of mutant G core promoters that contain 1-, 2-, or 3-nucleotide insertions or deletions between the DPE and Inr. wt, wild type. (B) DNase I footprint analysis of _G_−1, G wild-type, and G+1 core promoters with purified Drosophila TFIID. Arrows indicate DNase I hypersensitive sites.
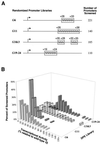
FIG. 3
Analysis of the range of sequences that can function as DPE motifs. (A) Diagram of randomized G core promoter libraries. Four promoter libraries were constructed with G core promoter sequences (−2 to +40 relative to the transcription start site), except that the portions of the sequence indicated by N's contained approximately equivalent amounts of each of the four deoxyribonucleotides. (B) Summary of the in vitro transcription screening of the randomized G core promoter libraries. Individual clones from each of the randomized libraries were isolated and then subjected to in vitro transcription analysis. The graph shows the distribution of transcriptional activity for each of the tested promoters relative to the wild-type G core promoter (100%) for each library.
FIG. 4
The DPE appears to be present in many Drosophila promoters. (A) The frequency of occurrence of the DPE appears to be comparable to that of the TATA box in Drosophila core promoters. A Drosophila core promoter database was created by aligning sequences of 205 Drosophila core promoters with accurately determined transcription start sites. The number of promoters that appear to possess a TATA box only, a DPE only, both elements, or neither element is shown. TATA boxes were defined as sequences with at least a 5 out of 6 match with the TATAAA sequence upstream of −20 relative to the transcription start site. DPE motifs were defined as sequences with at least a 5 out of 6 match with the DPE functional range set (Table 2) at exactly +28 to +33 relative to the start site. The Drosophila core promoter database is available at the website
http://www-biology.ucsd.edu/labs/Kadonaga/DCPD.html
. (B) Nucleotide distributions in the upstream region of Drosophila core promoters. The nucleotide distributions at positions −47 to −3 relative to the transcriptional start site (+1) were analyzed for 59 TATA-only promoters, 54 DPE-only promoters, 28 TATA + DPE promoters, and 64 TATA-less and DPE-less promoters. A 1242 test of the null hypothesis that each nucleotide is equally distributed was performed for every position. Letters over a bracket above the bars of the graph indicate the overrepresented nucleotides at positions that significantly deviate from the null hypothesis (P < 0.001). (C) Nucleotide distributions in the downstream region of Drosophila core promoters. The downstream region (from −2 to +45 relative to the start site) of Drosophila core promoters was analyzed as in panel B. The Inr and DPE motifs are indicated with a bracket below the graphs.
FIG. 4
The DPE appears to be present in many Drosophila promoters. (A) The frequency of occurrence of the DPE appears to be comparable to that of the TATA box in Drosophila core promoters. A Drosophila core promoter database was created by aligning sequences of 205 Drosophila core promoters with accurately determined transcription start sites. The number of promoters that appear to possess a TATA box only, a DPE only, both elements, or neither element is shown. TATA boxes were defined as sequences with at least a 5 out of 6 match with the TATAAA sequence upstream of −20 relative to the transcription start site. DPE motifs were defined as sequences with at least a 5 out of 6 match with the DPE functional range set (Table 2) at exactly +28 to +33 relative to the start site. The Drosophila core promoter database is available at the website
http://www-biology.ucsd.edu/labs/Kadonaga/DCPD.html
. (B) Nucleotide distributions in the upstream region of Drosophila core promoters. The nucleotide distributions at positions −47 to −3 relative to the transcriptional start site (+1) were analyzed for 59 TATA-only promoters, 54 DPE-only promoters, 28 TATA + DPE promoters, and 64 TATA-less and DPE-less promoters. A 1242 test of the null hypothesis that each nucleotide is equally distributed was performed for every position. Letters over a bracket above the bars of the graph indicate the overrepresented nucleotides at positions that significantly deviate from the null hypothesis (P < 0.001). (C) Nucleotide distributions in the downstream region of Drosophila core promoters. The downstream region (from −2 to +45 relative to the start site) of Drosophila core promoters was analyzed as in panel B. The Inr and DPE motifs are indicated with a bracket below the graphs.
FIG. 4
The DPE appears to be present in many Drosophila promoters. (A) The frequency of occurrence of the DPE appears to be comparable to that of the TATA box in Drosophila core promoters. A Drosophila core promoter database was created by aligning sequences of 205 Drosophila core promoters with accurately determined transcription start sites. The number of promoters that appear to possess a TATA box only, a DPE only, both elements, or neither element is shown. TATA boxes were defined as sequences with at least a 5 out of 6 match with the TATAAA sequence upstream of −20 relative to the transcription start site. DPE motifs were defined as sequences with at least a 5 out of 6 match with the DPE functional range set (Table 2) at exactly +28 to +33 relative to the start site. The Drosophila core promoter database is available at the website
http://www-biology.ucsd.edu/labs/Kadonaga/DCPD.html
. (B) Nucleotide distributions in the upstream region of Drosophila core promoters. The nucleotide distributions at positions −47 to −3 relative to the transcriptional start site (+1) were analyzed for 59 TATA-only promoters, 54 DPE-only promoters, 28 TATA + DPE promoters, and 64 TATA-less and DPE-less promoters. A 1242 test of the null hypothesis that each nucleotide is equally distributed was performed for every position. Letters over a bracket above the bars of the graph indicate the overrepresented nucleotides at positions that significantly deviate from the null hypothesis (P < 0.001). (C) Nucleotide distributions in the downstream region of Drosophila core promoters. The downstream region (from −2 to +45 relative to the start site) of Drosophila core promoters was analyzed as in panel B. The Inr and DPE motifs are indicated with a bracket below the graphs.
FIG. 5
The DPE functional range set identifies additional DPE-dependent promoters. (A) In vitro transcription analysis of the EF-1α F1 and Sodh-1 core promoters. The sequences of the wild-type (Wt) and DPE mutant (Mut) versions of the promoters are indicated. (B) Scanning clustered point mutational analysis of the EF-1α F1 core promoter. A series of mutant core promoters with triple nucleotide substitutions, as indicated, were constructed and subjected to in vitro transcription and primer extension analysis. (C) DNase I footprint analysis of the EF-1α F1 promoter with purified Drosophila TFIID. The DPE mutant version of the EF-1α F1 promoter is identical to that used in panel A.
FIG. 6
The +24 position contributes to DPE promoter function. The wild-type (Wt) and +24 mutant (Mut) versions of the indicated promoters were subjected to in vitro transcription and primer extension analysis.
FIG. 7
A model of two distinct interactions of TFIID with TATA- versus DPE-driven core promoters. The model is discussed in the text. TAFs, TBP-associated factors.
Similar articles
- Drosophila TFIID binds to a conserved downstream basal promoter element that is present in many TATA-box-deficient promoters.
Burke TW, Kadonaga JT. Burke TW, et al. Genes Dev. 1996 Mar 15;10(6):711-24. doi: 10.1101/gad.10.6.711. Genes Dev. 1996. PMID: 8598298 - The downstream core promoter element, DPE, is conserved from Drosophila to humans and is recognized by TAFII60 of Drosophila.
Burke TW, Kadonaga JT. Burke TW, et al. Genes Dev. 1997 Nov 15;11(22):3020-31. doi: 10.1101/gad.11.22.3020. Genes Dev. 1997. PMID: 9367984 Free PMC article. - Caudal, a key developmental regulator, is a DPE-specific transcriptional factor.
Juven-Gershon T, Hsu JY, Kadonaga JT. Juven-Gershon T, et al. Genes Dev. 2008 Oct 15;22(20):2823-30. doi: 10.1101/gad.1698108. Genes Dev. 2008. PMID: 18923080 Free PMC article. - The DPE, a conserved downstream core promoter element that is functionally analogous to the TATA box.
Burke TW, Willy PJ, Kutach AK, Butler JE, Kadonaga JT. Burke TW, et al. Cold Spring Harb Symp Quant Biol. 1998;63:75-82. doi: 10.1101/sqb.1998.63.75. Cold Spring Harb Symp Quant Biol. 1998. PMID: 10384272 Review. No abstract available. - The DPE, a core promoter element for transcription by RNA polymerase II.
Kadonaga JT. Kadonaga JT. Exp Mol Med. 2002 Sep 30;34(4):259-64. doi: 10.1038/emm.2002.36. Exp Mol Med. 2002. PMID: 12515390 Review.
Cited by
- Expression of nonclassical MHC class Ib genes: comparison of regulatory elements.
Howcroft T, Singer D. Howcroft T, et al. Immunol Res. 2003;27(1):1-30. doi: 10.1385/IR:27:1:1. Immunol Res. 2003. PMID: 12637766 Review. - Identification of virion-associated transcriptional transactivator (VATT) of SGIV ICP46 promoter and their binding site on promoter.
Xia LQ, Chen JL, Zhang HL, Cai J, Zhou S, Lu YS. Xia LQ, et al. Virol J. 2019 Sep 3;16(1):110. doi: 10.1186/s12985-019-1210-0. Virol J. 2019. PMID: 31481132 Free PMC article. - Cis- and trans-acting influences on telomeric position effect in Drosophila melanogaster detected with a subterminal transgene.
Mason JM, Konev AY, Golubovsky MD, Biessmann H. Mason JM, et al. Genetics. 2003 Mar;163(3):917-30. doi: 10.1093/genetics/163.3.917. Genetics. 2003. PMID: 12663532 Free PMC article. - Core promoter structure in the oomycete Phytophthora infestans.
McLeod A, Smart CD, Fry WE. McLeod A, et al. Eukaryot Cell. 2004 Feb;3(1):91-9. doi: 10.1128/EC.3.1.91-99.2004. Eukaryot Cell. 2004. PMID: 14871940 Free PMC article. - Promoter-proximal tethering elements regulate enhancer-promoter specificity in the Drosophila Antennapedia complex.
Calhoun VC, Stathopoulos A, Levine M. Calhoun VC, et al. Proc Natl Acad Sci U S A. 2002 Jul 9;99(14):9243-7. doi: 10.1073/pnas.142291299. Epub 2002 Jul 1. Proc Natl Acad Sci U S A. 2002. PMID: 12093913 Free PMC article.
References
- Berk A J. Activation of RNA polymerase II transcription. Curr Opin Cell Biol. 1999;11:330–335. - PubMed
- Bucher P. Weight matrix descriptions of four eukaryotic RNA polymerase II promoter elements derived from 502 unrelated promoter sequences. J Mol Biol. 1990;212:563–578. - PubMed
- Burke T W, Kadonaga J T. Drosophila TFIID binds to a conserved downstream basal promoter element that is present in many TATA-box-deficient promoters. Genes Dev. 1996;10:711–724. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases