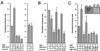Set domain-dependent regulation of transcriptional silencing and growth control by SUV39H1, a mammalian ortholog of Drosophila Su(var)3-9 - PubMed (original) (raw)
Set domain-dependent regulation of transcriptional silencing and growth control by SUV39H1, a mammalian ortholog of Drosophila Su(var)3-9
R Firestein et al. Mol Cell Biol. 2000 Jul.
Abstract
Mammalian SET domain-containing proteins define a distinctive class of chromatin-associated factors that are targets for growth control signals and oncogenic activation. SUV39H1, a mammalian ortholog of Drosophila Su(var)3-9, contains both SET and chromo domains, signature motifs for proteins that contribute to epigenetic control of gene expression through effects on the regional organization of chromatin structure. In this report we demonstrate that SUV39H1 represses transcription in a transient transcriptional assay when tethered to DNA through the GAL4 DNA binding domain. Under these conditions, SUV39H1 displays features of a long-range repressor capable of acting over several kilobases to silence basal promoters. A possible role in chromatin-mediated gene silencing is supported by the localization of exogenously expressed SUV39H1 to nuclear bodies with morphologic features suggestive of heterochromatin in interphase cells. In addition, we show that SUV39H1 is phosphorylated specifically at the G(1)/S cell cycle transition and when forcibly expressed suppresses cell growth. Growth suppression as well as the ability of SUV39H1 to form nuclear bodies and silence transcription are antagonized by the oncogenic antiphosphatase Sbf1 that when hyperexpressed interacts with the SET domain and stabilizes the phosphorylated form of SUV39H1. These studies suggest a phosphorylation-dependent mechanism for regulating the chromatin organizing activity of a mammalian su(var) protein and implicate the SET domain as a gatekeeper motif that integrates upstream signaling pathways to epigenetic regulation and growth control.
Figures
FIG. 1
Conservation and expression of SUV39H1, a mammalian ortholog of Su(var)3-9. Schematic depictions of the predicted protein compositions for human SUV39H1 and the orthologous Drosophila Su(var)3-9 and S. pombe CLR4 indicate the conserved chromo domains (light stipple), cysteine-rich regions (black box), SET domains (heavy stipple), and putative nuclear localization sequence (NLS). The portion of SUV39H1 used as immunogen for production of MAbs (α SUV) is shown below. SUV39H1SET is an N-terminal deletion mutant that contains an engineered N-terminal NLS. SUV39H1ΔSET and SUV39H1ΔC are C-terminal deletion mutants used in this study.
FIG. 2
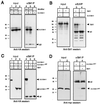
SUV39H1 displays SET domain-dependent physical association with the anti-phosphatase Sbf1. (A) 293t cells were cotransfected with expression constructs encoding HA-tagged SUV39H1, Sbf1, and Sbf1HCS as indicated above the gel lanes. Whole cell extracts prepared 48 h after transfection were subjected to immunoprecipitation (IP) using an anti-Sbf1 MAb. Detection of coprecipitating SUV39H1 by Western blot analysis using an anti-HA antibody demonstrated that it was capable of associating with both Sbf1 and Sbf1HCS. (B) Lysates of 293t cells transfected with constructs expressing the proteins indicated above the gel lanes were subjected to immunoprecipitation using an anti-SUV39H1 antibody. Coprecipitating Sbf1 was detected by Western blot analysis using an anti-Sbf1 MAb. (C and D) Lysates of 293t cells transfected with tagged constructs expressing the proteins indicated above the gel lanes were subjected to immunoprecipitation using an anti-Sbf1 MAb. Coprecipitating SUV39H1 proteins were detected by Western blot analysis with an anti-HA or anti-Myc antibody. The anti-rat secondary antibody (A and C) cross-reacted with mouse IgG heavy chain used in the immunoprecipitations. The amount of lysate in each input lane (input) is equivalent to 2% of the amount applied to beads (IP). Protein migrations are indicated by arrows; sizes are indicated in kilodaltons.
FIG. 3
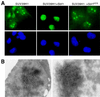
SUV39H1 forms nuclear bodies in vivo that are dispersed by Sbf1. (A) COS7 cells were examined by immunofluorescence 48 h after cotransfection of constructs expressing FLAG-tagged SUV39H1 in the presence or absence of a 10-fold excess of expression constructs for Sbf1 or Sbf1HCS. Green fluorescence corresponds to FLAG-tagged SUV39H1 staining which was revealed using primary anti-FLAG and secondary fluorescein isothiocyanate-conjugated antibodies. DAPI staining is shown in blue. Expression of transfected Sbf1 and Sbf1HCS was comparable as detected by Western blot analysis (data not shown). Magnification, ×630. (B) 293t cells were analyzed by immunoelectron microscopy 48 h after transfection with a construct expressing FLAG-tagged SUV39H1. Immune complexes were visualized using a primary antibody directed against the FLAG epitope tag and a secondary goat anti-mouse IgG conjugated with colloidal gold. Magnification, ×42,300.
FIG. 4
SUV39H1 displays transcriptional repressor properties that are modulated by Sbf1. (A) Expression constructs coding for the GAL4 DBD itself or a GAL4-SUV39H1 fusion protein (DBD-SUV) were cotransfected into COS7 cells in combination with a luciferase reporter gene under control of the myelomonocytic growth factor promoter. The amount (micrograms) of each construct present in the transfections is indicated below the histograms. Transcriptional activation is expressed as normalized luciferase units that have been corrected for β-galactosidase expression from an internal control lacZ construct in each transfection. The data represent the means from at least three independent experiments. Transcriptional repression observed for GAL4-SUV was dependent on the presence of GAL4 binding sites in the reporter gene and not observed if SUV39H1 was untethered to the GAL4 DBD (not shown). (B) Transcriptional assays were conducted as described for panel A except that the luciferase reporter gene constructs contained a minimal SV40 promoter separated by variable distances (indicated below histograms) from upstream GAL4 DNA binding sites. (C) Transcriptional assays were performed as described for panel A with the addition of expression constructs encoding Sbf1 (amino acids 700 to 1931) or Sbf1HCS (as indicated below the histograms) at fivefold excess concentration compared to cotransfected SUV39H1 constructs. Repression of the myelomonocytic growth factor promoter by GAL4-SUV was partially alleviated by coexpressed Sbf1 but not Sbf1HCS. Repression was also observed by GAL4-SUVΔC but was not relieved by coexpressed Sbf1. Western blots demonstrating comparable expression levels of transfected Sbf1 and Sbf1HCS as well as GAL4-SUV and GAL4-SUVΔC are shown as insets.
FIG. 5
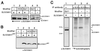
SUV39H1 undergoes SET-dependent phosphorylation that is enhanced by Sbf1. (A) Bosc cells were transduced with retroviral vectors coexpressing Sbf1 or Sbf1HCS (from an IRES element) with SUV39H1. Cells were harvested 2 days after transduction, and equal amounts of whole cell lysate used for Western blotting. Shifted (pSUV39H1) and nonshifted (SUV39H1) forms of SUV39H1 were detected using an anti-SUV39H1 antibody. Similar shifts in the migration of exogenous (lane 3) or endogenous (lane 4) SUV39H1 were induced by forced expression of Sbf1. Expression of transfected Sbf1 and Sbf1HCS was comparable as detected by Western blot analysis (data not shown). (B) Analyses similar to those in panel A, substituting SUV39H1ΔSET for SUV39H1, showed no shifted migration of SUV39H1ΔSET following coexpression with Sbf1. (C) HeLa cells transfected with control or SUV39H1-expressing vectors were metabolically labeled with [32P]orthophosphate. Equal amounts of nuclear extracts were immunoprecipitated (IP) using anti-SUV39H1 or anti-Pbx1 (nonimmune) antibodies. Precipitated proteins were fractionated by SDS-PAGE and subjected to autoradiography. In parallel on the same gel, lysate from cells cotransfected with Sbf1 and SUV39H1 was analyzed by Western blotting to determine the migration of shifted (pSUV39H1) and nonshifted (SUV39H1) forms of SUV39H1.
FIG. 6
SUV39H1 is phosphorylated at the transition from G1 to S phase of the cell cycle. HeLa cells were growth arrested by serum starvation for 48 h in tissue culture medium. Cells were then stimulated to synchronously reenter the cell cycle by addition of serum-rich medium. Protein lysates were prepared from nonstimulated cells (0) and at hourly time points (indicated above the gel lanes) following serum stimulation. Endogenous SUV39H1 proteins were detected by Western blotting using an anti-SUV39H1 MAb. Migrations of hypo- and hyperphosphorylated SUV39H1 proteins are indicated. The entry of cells into S phase was determined by measuring BrdU incorporation (indicated by + or − below the panel) in parallel cultures following 30-min BrdU pulse-labeling.
FIG. 7
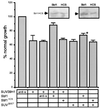
SUV39H1 has growth-inhibitory properties that are reversed by Sbf1. NIH 3T3 cells were transduced with retroviral stocks expressing SUV39H1 alone or in combination with GFP, Sbf1 (amino acids 1091 to 1861), or Sbf1HCS (indicated below histogram), using an IRES element. SUV39H1 and Sbf1 protein expression in transduced cells was confirmed by Western blotting using anti-SUV39H1 and Sbf1 antibodies. Growth rates were determined by measuring BrdU incorporation in equal numbers of transduced NIH 3T3 cells that were plated 24 h previously. Cells staining positively for BrdU incorporation were counted as a fraction of cells that expressed GFP (growth fraction) or total cells. The growth fraction of cells infected with GFP alone was arbitrarily set at 100%, and percent growth rate was calculated accordingly. Western blots showing expression levels of exogenous Sbf1 and Sbf1HCS are shown as insets above their corresponding panels. Presented data represent the means and standard deviations from three separate experiments. anti-s, cDNA insert in reverse orientation; ∗, growth fraction was not significantly different from SUVΔSET alone (P > 0.05).
Similar articles
- Functional mammalian homologues of the Drosophila PEV-modifier Su(var)3-9 encode centromere-associated proteins which complex with the heterochromatin component M31.
Aagaard L, Laible G, Selenko P, Schmid M, Dorn R, Schotta G, Kuhfittig S, Wolf A, Lebersorger A, Singh PB, Reuter G, Jenuwein T. Aagaard L, et al. EMBO J. 1999 Apr 1;18(7):1923-38. doi: 10.1093/emboj/18.7.1923. EMBO J. 1999. PMID: 10202156 Free PMC article. - Structure-function analysis of SUV39H1 reveals a dominant role in heterochromatin organization, chromosome segregation, and mitotic progression.
Melcher M, Schmid M, Aagaard L, Selenko P, Laible G, Jenuwein T. Melcher M, et al. Mol Cell Biol. 2000 May;20(10):3728-41. doi: 10.1128/MCB.20.10.3728-3741.2000. Mol Cell Biol. 2000. PMID: 10779362 Free PMC article. - Mitotic phosphorylation of SUV39H1, a novel component of active centromeres, coincides with transient accumulation at mammalian centromeres.
Aagaard L, Schmid M, Warburton P, Jenuwein T. Aagaard L, et al. J Cell Sci. 2000 Mar;113 ( Pt 5):817-29. doi: 10.1242/jcs.113.5.817. J Cell Sci. 2000. PMID: 10671371 - Histone modification and the control of heterochromatic gene silencing in Drosophila.
Ebert A, Lein S, Schotta G, Reuter G. Ebert A, et al. Chromosome Res. 2006;14(4):377-92. doi: 10.1007/s10577-006-1066-1. Chromosome Res. 2006. PMID: 16821134 Review. - Mammalian Su(var) genes in chromatin control.
Fodor BD, Shukeir N, Reuter G, Jenuwein T. Fodor BD, et al. Annu Rev Cell Dev Biol. 2010;26:471-501. doi: 10.1146/annurev.cellbio.042308.113225. Annu Rev Cell Dev Biol. 2010. PMID: 19575672 Review.
Cited by
- p38α MAPK disables KMT1A-mediated repression of myogenic differentiation program.
Chatterjee B, Wolff DW, Jothi M, Mal M, Mal AK. Chatterjee B, et al. Skelet Muscle. 2016 Aug 22;6:28. doi: 10.1186/s13395-016-0100-z. eCollection 2016. Skelet Muscle. 2016. PMID: 27551368 Free PMC article. - Relationship between histone H3 lysine 9 methylation, transcription repression, and heterochromatin protein 1 recruitment.
Stewart MD, Li J, Wong J. Stewart MD, et al. Mol Cell Biol. 2005 Apr;25(7):2525-38. doi: 10.1128/MCB.25.7.2525-2538.2005. Mol Cell Biol. 2005. PMID: 15767660 Free PMC article. - The p75NTR-interacting protein SC1 inhibits cell cycle progression by transcriptional repression of cyclin E.
Chittka A, Arevalo JC, Rodriguez-Guzman M, Pérez P, Chao MV, Sendtner M. Chittka A, et al. J Cell Biol. 2004 Mar 29;164(7):985-96. doi: 10.1083/jcb.200301106. J Cell Biol. 2004. PMID: 15051733 Free PMC article. - Central role of Drosophila SU(VAR)3-9 in histone H3-K9 methylation and heterochromatic gene silencing.
Schotta G, Ebert A, Krauss V, Fischer A, Hoffmann J, Rea S, Jenuwein T, Dorn R, Reuter G. Schotta G, et al. EMBO J. 2002 Mar 1;21(5):1121-31. doi: 10.1093/emboj/21.5.1121. EMBO J. 2002. PMID: 11867540 Free PMC article. - CDK2-dependent phosphorylation of Suv39H1 is involved in control of heterochromatin replication during cell cycle progression.
Park SH, Yu SE, Chai YG, Jang YK. Park SH, et al. Nucleic Acids Res. 2014 Jun;42(10):6196-207. doi: 10.1093/nar/gku263. Epub 2014 Apr 11. Nucleic Acids Res. 2014. PMID: 24728993 Free PMC article.
References
- Aagaard L, Laible G, Selenko P, Schmid M, Dorn R, Schotta G, Kuhfittig S, Wolf A, Lebersorger A S, Reuter G, Jenuwein T. Functional mammalian homologues of the drosophila PEV-modifier Su(var)3-9 encode centromere-associated proteins which complex with the heterochromatin component M31. EMBO J. 1999;18:1923–1938. - PMC - PubMed
- Baksa K, Morawietz H, Dombradi V, Axton M, Taubert H, Szabo G, Torok I, Udvardy A, Gyurkovics H, Szoor B. Mutations in the protein phosphatase I gene at 87B can differentially affect suppression of position-effect variegation and mitosis in Drosophila melanogaster. Genetics. 1993;135:117–125. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases