Disruption of nuclear lamin organization blocks the elongation phase of DNA replication - PubMed (original) (raw)
Disruption of nuclear lamin organization blocks the elongation phase of DNA replication
R D Moir et al. J Cell Biol. 2000.
Abstract
The role of nuclear lamins in DNA replication is unclear. To address this, nuclei were assembled in Xenopus extracts containing AraC, a reversible inhibitor that blocks near the onset of the elongation phase of replication. Dominant-negative lamin mutants lacking their NH(2)-terminal domains were added to assembled nuclei to disrupt lamin organization. This prevented the resumption of DNA replication after the release of the AraC block. This inhibition of replication was not due to gross disruption of nuclear envelope structure and function. The organization of initiation factors was not altered by lamin disruption, and nuclei resumed replication when transferred to extracts treated with CIP, an inhibitor of the cyclin-dependent kinase (cdk) 2-dependent step of initiation. This suggests that alteration of lamin organization does not affect the initiation phase of DNA replication. Instead, we find that disruption of lamin organization inhibited chain elongation in a dose-dependent fashion. Furthermore, the established organization of two elongation factors, proliferating cell nuclear antigen, and replication factor complex, was disrupted by DeltaNLA. These findings demonstrate that lamin organization must be maintained in nuclei for the elongation phase of DNA replication to proceed.
Figures
Figure 1
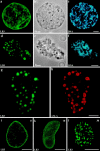
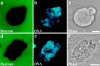
ΔNLA disrupts lamin organization in AraC-arrested nuclei. a–c depict confocal sections from a nucleus exposed to protein buffer, and d–f show a nucleus exposed to ΔNLA. In both cases, nuclei were assembled in interphase extract containing 0.2 mM AraC for 70 min before the addition of an equal volume of ΔNLA or protein buffer, and 90 min later the nuclei were fixed. The control nucleus stained with a mAb directed against lamin B3 (a) reveals a bright rim pattern accompanied by nucleoplasmic staining. In the ΔNLA-disrupted nucleus (d), the LB3 staining is absent from the rim and is instead found in abnormal nucleoplasmic aggregates. These aggregates are clearly visible with phase optics (e). The DNA staining patterns (Hoechst dye) of both the control (c) and disrupted nuclei (f) are also shown. Immunofluorescence using a mAb directed against lamin B3 (g) and a polyclonal antibody directed against human lamin A (h) shows that the ΔNLA protein colocalizes with the endogenous LB3 in nucleoplasmic aggregates. In larger aggregates, the mutant lamin appears to be more concentrated at the periphery, whereas the endogenous lamin is concentrated toward the center. The addition of ΔNLA produced a rapid alteration of lamin organization. 10 min after the addition of the mutant protein, the lamina staining is less apparent and a fine punctate pattern of LB3 staining appears through the nucleus (compare i with k). 30 min after the addition of the mutant protein, LB3 is found in abnormal nucleoplasmic aggregates (l). Each image represents a single optical section through the central region of a nucleus. Bars: 5 μm (a–h); and 10 μm (i–l).
Figure 2
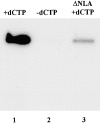
Autoradiograph demonstrating that lamin disruption blocks the resumption of DNA replication in AraC-arrested nuclei. Nuclei were assembled in an interphase extract containing AraC. In lane 1, dCTP was added with [32P]dATP to relieve the AraC block of replication. In lane 2, the dCTP was omitted. In lane 3, dCTP and [32P]dATP were added to AraC-arrested nuclei after disruption by ΔNLA. For all samples, nuclei were allowed to incorporate [32P]dATP for 90 min. The samples were resolved on a 1% TBE agarose gel, and the dried gel was used for autoradiography. Quantitation with a beta imager revealed that the addition of ΔNLA reduced the incorporation of [32P]dATP by >95% (not shown).
Figure 3
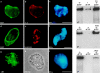
Lamin organization and DNA replication were unaltered by the addition of LA or NLS-vimentin. However, the addition of ΔNLB3 disrupted lamin organization and blocked DNA replication. Each of the bacterially expressed proteins was added to nuclei that had been assembled in extract containing AraC for 70 min. When LA is added, the LB3 staining pattern appears undisrupted (a) and the LA staining pattern was indistinguishable from that of LB3 as shown by immunofluorescence with a lamin A–specific antibody (b). Furthermore, the addition of LA did not prevent the resumption of replication that occurs with the addition of dCTP in DNA replication assays (c*, compare first and third lanes). The addition of NLS-vimentin also does not disrupt the LB3 staining (d), although the NLS-vimentin appears in large aggregates as shown by staining with a vimentin antibody (e). The addition of NLS-vimentin did not prevent the resumption of DNA replication (f*, compare first and third lanes). The addition of the ΔNLB3 disrupts the endogenous lamin network (g), resulting in large nucleoplasmic aggregates seen in immunofluorescence (g) and phase (h). The presence of ΔNLB3 prevents the resumption of DNA replication (i*, compare first and third lanes). All images were obtained using a Zeiss axiovert microscope. Bars, 10 μm.
Figure 4
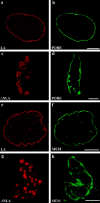
Lamin disruption does not obviously alter the morphology of the nuclear membrane or pores. Control nuclei (treated with LA) and lamin-disrupted nuclei (treated with ΔNLA) were stained with 414 (b and d), a mAb that reacts with the nucleoporins or with the membrane dye DIOC6 (f and h). Both the pore and membrane markers produce a bright rim stain that aligns with the edge of nuclei. The LA (a and e) and ΔNLA (c and g) were stained with an antibody directed against human lamin A. The lamin aggregates did not stain with either the pore (c and d) or membrane marker (g and h). Each pair of confocal images represents an optical section through the central region of a nucleus. Bars, 5 μm.
Figure 5
To assess the barrier function of the lamin-disrupted nuclei, FITC-labeled dextran (150 kD) was added to extracts containing normal (a–c) or ΔNLA-disrupted (d–e) nuclei. Aliquots of each reaction were transferred to slides and observed by epifluorescence. Nuclei were located using Hoechst dye (b and e), and observed in the FITC channel (a and d). Both the control and disrupted nuclei appear dark in the FITC channel, demonstrating that both types of nuclei exclude the FITC-labeled dextran. The phase contrast images of these nuclei are seen in c and f. The disrupted nucleus (f) contains phase-dense bodies that represent the lamin aggregates. Over 95% of both control and disrupted nuclei excluded the dextran (n = 100). Bars, 10 μm.
Figure 7
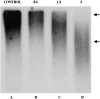
ΔNLA blocks the synthesis of high molecular weight replication products. Increasing concentrations of ΔNLA (0, 0.6, 1.5, and 3 μM) were added to extracts containing AraC-arrested nuclei. After lamin disruption, dCTP was added to each reaction to release the AraC block and replication was allowed to proceed for 10 min. Subsequently [32P]dATP was added to the reactions, and 2 min later the reactions were stopped. The replication products were resolved on a denaturing alkaline agarose gel, and the dried gel was used for autoradiography. The average size of the replication products synthesized in the reaction lacking ΔNLA (lane A) was >10,000 bases. As the concentration of ΔNLA added was increased, the size of the replication products decreased (lanes B–D), such that at 3 μM (lane D), the average size was ∼1,500 bases. The arrows on the right are size markers indicating the approximate position of markers 2,027 and 9,416 bases in length (New England Biolabs, Inc.).
Figure 6
To assess nuclear transport in disrupted nuclei, TRITC-labeled human serum albumin (HSA) coupled to an NLS peptide (see Materials and Methods) was added to extracts containing ΔNLA-disrupted (a–d) and control (untreated, e–h) nuclei. 30 min later, the preparations were fixed and a portion of each was observed by conventional epifluorescence. Nuclei were located using a DNA dye and scored as positive for transport if they also fluoresced in the TRITC channel. A lamin-disrupted nucleus (a and b) and a control nucleus (e and f) are shown. A comparable fraction of nuclei from both reactions imported the labeled HSA over the 30-min time period (a and c, 81% control, 83% ΔNLA; n = 85 for each preparation). There was no detectable difference in the range of fluorescence intensities of the transporting nuclei between these two samples. To ensure the mutant lamin efficiently disrupted lamin organization, a sample of the transport reaction was fixed and stained for the endogenous LB3 distribution. More than 98% of nuclei treated with ΔNLA had disrupted lamin organization compared with control samples (compare d with h). Bars, 10 μm.
Figure 8
ΔNLA does not disrupt the cdk2-dependent initiation step of replication. Nuclei were assembled in the presence of AraC and subsequently, the protein buffer control (a) or ΔNLA (b) were added. After an additional 90-min incubation, the nuclei were transferred to extract containing AraC and either CIP, ΔNLA, or protein buffer. Replication was monitored with the addition of dCTP and [32P]dATP, and after a 60-min labeling period the reactions were stopped and the replication products resolved on a nondenaturing agarose gel. The image is an autoradiograph of the dry gel. (a) The control nuclei resume replication when transferred to extract containing buffer (lane 1) or CIP (lane 3) as compared with the sample in which the AraC block was not reversed (lane 2). (b) Similarly, ΔNLA-disrupted nuclei resume replication when transferred to extract containing buffer (lane 4) or CIP (lane 6). A sample in which the AraC block was not reversed is shown in lane 5. If the disrupted nuclei were transferred to an extract that also contained ΔNLA, then replication did not resume (lane 7).
Figure 9
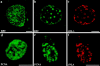
Disruption of lamin organization alters the distributions of RFC and PCNA. Double immunofluorescence of nuclei assembled in the presence of AraC and subsequently treated with buffer or ΔNLA. The nuclei were stained with a mAb directed against the large subunit of RFC (a and b) or PCNA (d and e) and an antibody directed against ΔNLA (c and f). The AraC-arrested control nuclei display normal RFC (a) and PCNA (d) staining patterns, i.e., relatively diffuse staining spread throughout the nuclei. In contrast, both RFC (b) and PCNA (e) are found in abnormal aggregates in the lamin-disrupted nuclei. The mutant lamin is also found associated with these abnormal aggregates (c and f). The size of the aggregates varied slightly between experiments. In cases in which large aggregates are produced (i.e., b) the RFC and PCNA are found in the center of the aggregate with ΔNLA concentrated at the edge of the aggregate. Images represent single confocal optical sections through the central region of a nucleus. Bars, 10 μm.
Figure 10
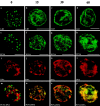
Normal distributions of nuclear lamins and PCNA are rapidly reestablished when lamin-disrupted nuclei are transferred to an untreated Xenopus extract. Nuclei were assembled in the presence of AraC and subsequently lamin organization was disrupted by the addition of ΔNLA. The disrupted nuclei were transferred to an untreated extract and fixed at the indicated timepoints (Materials and Methods). Parallel samples were stained for LB3 (a–d) or PCNA and DNA (e–p). The samples in the far left column were fixed before transfer (0 time). At 0 time, LB3 was found in the characteristic nucleoplasmic aggregates (a), but 15 min after transfer to untreated extract, the larger lamin aggregates are no longer visible (b). Instead, a more normal appearance with rim staining and finer speckled nucleoplasmic lamin standing was observed. A nearly normal lamin morphology was apparent 60 min after the transfer (d). Similarly, the PCNA distribution rapidly changed after transfer to untreated extract (e–p). In disrupted nuclei, PCNA appears in large aggregates that do not coalign with chromatin. As early as 15 min after transfer, PCNA staining was no longer found in these aggregates, but instead regained its normal distribution as a finely speckled pattern along the chromatin (f, j, and n). This colocalization with chromatin remained evident 60 min after transfer (h, l, and p). Bars, 10 μm.
Similar articles
- Cip1 blocks the initiation of DNA replication in Xenopus extracts by inhibition of cyclin-dependent kinases.
Strausfeld UP, Howell M, Rempel R, Maller JL, Hunt T, Blow JJ. Strausfeld UP, et al. Curr Biol. 1994 Oct 1;4(10):876-83. doi: 10.1016/s0960-9822(00)00196-2. Curr Biol. 1994. PMID: 7850420 - Disruption of nuclear lamin organization alters the distribution of replication factors and inhibits DNA synthesis.
Spann TP, Moir RD, Goldman AE, Stick R, Goldman RD. Spann TP, et al. J Cell Biol. 1997 Mar 24;136(6):1201-12. doi: 10.1083/jcb.136.6.1201. J Cell Biol. 1997. PMID: 9087437 Free PMC article. - Separate domains of p21 involved in the inhibition of Cdk kinase and PCNA.
Chen J, Jackson PK, Kirschner MW, Dutta A. Chen J, et al. Nature. 1995 Mar 23;374(6520):386-8. doi: 10.1038/374386a0. Nature. 1995. PMID: 7885482 - Early events in DNA replication require cyclin E and are blocked by p21CIP1.
Jackson PK, Chevalier S, Philippe M, Kirschner MW. Jackson PK, et al. J Cell Biol. 1995 Aug;130(4):755-69. doi: 10.1083/jcb.130.4.755. J Cell Biol. 1995. PMID: 7642695 Free PMC article. - Xenopus lamin B3 has a direct role in the assembly of a replication competent nucleus: evidence from cell-free egg extracts.
Goldberg M, Jenkins H, Allen T, Whitfield WG, Hutchison CJ. Goldberg M, et al. J Cell Sci. 1995 Nov;108 ( Pt 11):3451-61. doi: 10.1242/jcs.108.11.3451. J Cell Sci. 1995. PMID: 8586657
Cited by
- Lamin A/C facilitates DNA damage response by modulating ATM signaling and homologous recombination pathways.
Kim SJ, Park SH, Myung K, Lee KY. Kim SJ, et al. Anim Cells Syst (Seoul). 2024 Aug 20;28(1):401-416. doi: 10.1080/19768354.2024.2393820. eCollection 2024. Anim Cells Syst (Seoul). 2024. PMID: 39176289 Free PMC article. - The impact of tumor microenvironment: unraveling the role of physical cues in breast cancer progression.
Akinpelu A, Akinsipe T, Avila LA, Arnold RD, Mistriotis P. Akinpelu A, et al. Cancer Metastasis Rev. 2024 Jun;43(2):823-844. doi: 10.1007/s10555-024-10166-x. Epub 2024 Jan 19. Cancer Metastasis Rev. 2024. PMID: 38238542 Free PMC article. Review. - Integration of nuclear Ca2+ transients and subnuclear protein shuttling provides a novel mechanism for the regulation of CREB-dependent gene expression.
Karpova A, Samer S, Turacak R, Yuanxiang P, Kreutz MR. Karpova A, et al. Cell Mol Life Sci. 2023 Jul 25;80(8):228. doi: 10.1007/s00018-023-04876-8. Cell Mol Life Sci. 2023. PMID: 37491479 Free PMC article. - Loss of FAM111B protease mutated in hereditary fibrosing poikiloderma negatively regulates telomere length.
Kliszczak M, Moralli D, Jankowska JD, Bryjka P, Subha Meem L, Goncalves T, Hester SS, Fischer R, Clynes D, Green CM. Kliszczak M, et al. Front Cell Dev Biol. 2023 Jun 5;11:1175069. doi: 10.3389/fcell.2023.1175069. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 37342232 Free PMC article. - Nuclear envelope, chromatin organizers, histones, and DNA: The many achilles heels exploited across cancers.
Balaji AK, Saha S, Deshpande S, Poola D, Sengupta K. Balaji AK, et al. Front Cell Dev Biol. 2022 Dec 16;10:1068347. doi: 10.3389/fcell.2022.1068347. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36589746 Free PMC article. Review.
References
- Aebi U., Cohn J., Buhle L., Gerace L. The nuclear lamina is a meshwork of intermediate-type filaments. Nature. 1986;323:560–564. - PubMed
- Blow J.J., Laskey R.A. A role for the nuclear envelope in controlling DNA replication within the cell cycle. Nature. 1988;332:546–548. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous