Purification and characterization of a membrane-bound hydrogenase from the hyperthermophilic archaeon Pyrococcus furiosus - PubMed (original) (raw)
Purification and characterization of a membrane-bound hydrogenase from the hyperthermophilic archaeon Pyrococcus furiosus
R Sapra et al. J Bacteriol. 2000 Jun.
Abstract
Highly washed membrane preparations from cells of the hyperthermophilic archaeon Pyrococcus furiosus contain high hydrogenase activity (9.4 micromol of H(2) evolved/mg at 80 degrees C) using reduced methyl viologen as the electron donor. The enzyme was solubilized with n-dodecyl-beta-D-maltoside and purified by multistep chromatography in the presence of Triton X-100. The purified preparation contained two major proteins (alpha and beta) in an approximate 1:1 ratio with a minimum molecular mass near 65 kDa and contained approximately 1 Ni and 4 Fe atoms/mol. The reduced enzyme gave rise to an electron paramagnetic resonance signal typical of the so-called Ni-C center of mesophilic NiFe-hydrogenases. Neither highly washed membranes nor the purified enzyme used NAD(P)(H) or P. furiosus ferredoxin as an electron carrier, nor did either catalyze the reduction of elemental sulfur with H(2) as the electron donor. Using N-terminal amino acid sequence information, the genes proposed to encode the alpha and beta subunits were located in the genome database within a putative 14-gene operon (termed mbh). The deduced sequences of the two subunits (Mbh 11 and 12) were distinctly different from those of the four subunits that comprise each of the two cytoplasmic NiFe-hydrogenases of P. furiosus and show that the alpha subunit contains the NiFe-catalytic site. Six of the open reading frames (ORFs) in the operon, including those encoding the alpha and beta subunits, show high sequence similarity (>30% identity) with proteins associated with the membrane-bound NiFe-hydrogenase complexes from Methanosarcina barkeri, Escherichia coli, and Rhodospirillum rubrum. The remaining eight ORFs encode small (<19-kDa) hypothetical proteins. These data suggest that P. furiosus, which was thought to be solely a fermentative organism, may contain a previously unrecognized respiratory system in which H(2) metabolism is coupled to energy conservation.
Figures
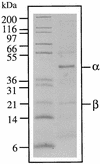
FIG. 1
SDS-polyacrylamide gel of the purified MB hydrogenase of P. furiosus. Marker proteins with the indicated molecular masses are in the left lane, and the purified hydrogenase is in the right lane. The two subunits of the hydrogenase (α and β) are indicated.
FIG. 2
Proposed operon encoding the MB hydrogenase complex of P. furiosus. The horizontal arrows near Mbh 11 and Mbh 12 indicate positions of the N-terminal amino acid sequences obtained from the purified complex. Also shown are the gene arrangements of the hyc, ech, and coo operons from E. coli, M. barkeri, and R. rubrum, respectively (8, 13, 18). Genes showing significant sequence similarity have the same shading.
Similar articles
- Intact functional fourteen-subunit respiratory membrane-bound [NiFe]-hydrogenase complex of the hyperthermophilic archaeon Pyrococcus furiosus.
McTernan PM, Chandrayan SK, Wu CH, Vaccaro BJ, Lancaster WA, Yang Q, Fu D, Hura GL, Tainer JA, Adams MW. McTernan PM, et al. J Biol Chem. 2014 Jul 11;289(28):19364-72. doi: 10.1074/jbc.M114.567255. Epub 2014 May 23. J Biol Chem. 2014. PMID: 24860091 Free PMC article. - Enzymes of hydrogen metabolism in Pyrococcus furiosus.
Silva PJ, van den Ban EC, Wassink H, Haaker H, de Castro B, Robb FT, Hagen WR. Silva PJ, et al. Eur J Biochem. 2000 Nov;267(22):6541-51. doi: 10.1046/j.1432-1327.2000.01745.x. Eur J Biochem. 2000. PMID: 11054105 - Engineering the respiratory membrane-bound hydrogenase of the hyperthermophilic archaeon Pyrococcus furiosus and characterization of the catalytically active cytoplasmic subcomplex.
McTernan PM, Chandrayan SK, Wu CH, Vaccaro BJ, Lancaster WA, Adams MW. McTernan PM, et al. Protein Eng Des Sel. 2015 Jan;28(1):1-8. doi: 10.1093/protein/gzu051. Epub 2014 Dec 3. Protein Eng Des Sel. 2015. PMID: 25476267 Free PMC article. - The modular respiratory complexes involved in hydrogen and sulfur metabolism by heterotrophic hyperthermophilic archaea and their evolutionary implications.
Schut GJ, Boyd ES, Peters JW, Adams MW. Schut GJ, et al. FEMS Microbiol Rev. 2013 Mar;37(2):182-203. doi: 10.1111/j.1574-6976.2012.00346.x. Epub 2012 Jul 12. FEMS Microbiol Rev. 2013. PMID: 22713092 Review. - Production and Application of a Soluble Hydrogenase from Pyrococcus furiosus.
Wu CH, McTernan PM, Walter ME, Adams MW. Wu CH, et al. Archaea. 2015 Oct 12;2015:912582. doi: 10.1155/2015/912582. eCollection 2015. Archaea. 2015. PMID: 26543406 Free PMC article. Review.
Cited by
- Identification of a novel class of membrane-bound [NiFe]-hydrogenases in Thermococcus onnurineus NA1 by in silico analysis.
Lim JK, Kang SG, Lebedinsky AV, Lee JH, Lee HS. Lim JK, et al. Appl Environ Microbiol. 2010 Sep;76(18):6286-9. doi: 10.1128/AEM.00123-10. Epub 2010 Jul 23. Appl Environ Microbiol. 2010. PMID: 20656864 Free PMC article. - Posttranslational protein modification in Archaea.
Eichler J, Adams MW. Eichler J, et al. Microbiol Mol Biol Rev. 2005 Sep;69(3):393-425. doi: 10.1128/MMBR.69.3.393-425.2005. Microbiol Mol Biol Rev. 2005. PMID: 16148304 Free PMC article. Review. - Novel multiprotein complexes identified in the hyperthermophilic archaeon Pyrococcus furiosus by non-denaturing fractionation of the native proteome.
Menon AL, Poole FL 2nd, Cvetkovic A, Trauger SA, Kalisiak E, Scott JW, Shanmukh S, Praissman J, Jenney FE Jr, Wikoff WR, Apon JV, Siuzdak G, Adams MW. Menon AL, et al. Mol Cell Proteomics. 2009 Apr;8(4):735-51. doi: 10.1074/mcp.M800246-MCP200. Epub 2008 Nov 28. Mol Cell Proteomics. 2009. PMID: 19043064 Free PMC article. - Formate addition enhanced hydrogen production by Thermococcus paralvinellae when grown on brewery wastewater.
Sistu H, Holden JF. Sistu H, et al. Front Microbiol. 2025 Mar 18;16:1560780. doi: 10.3389/fmicb.2025.1560780. eCollection 2025. Front Microbiol. 2025. PMID: 40170917 Free PMC article. - Energy-converting [NiFe] hydrogenases from archaea and extremophiles: ancestors of complex I.
Hedderich R. Hedderich R. J Bioenerg Biomembr. 2004 Feb;36(1):65-75. doi: 10.1023/b:jobb.0000019599.43969.33. J Bioenerg Biomembr. 2004. PMID: 15168611 Review.
References
- Adams M W W. The structure and mechanism of iron-hydrogenases. Biochim Biophys Acta. 1990;1020:115–145. - PubMed
- Adams M W W, Kletzin A. Oxidoreductase-type enzymes and redox proteins involved in fermentative metabolisms of hyperthermophilic archaea. Adv Protein Chem. 1996;48:101–180. - PubMed
- Adams M W W, Mortenson L E. The physical and catalytic properties of hydrogenase II of Clostridium pasteurianum. J Biol Chem. 1984;259:7045–7055. - PubMed
- Albracht S P J. Intimate relationships of the large and the small subunits of all nickel hydrogenases with two nuclear-encoded subunits of mitochondrial NADH: ubiquinone oxidoreductase. Biochim Biophys Acta. 1993;1144:221–224. - PubMed
- Albracht S P J. Nickel hydrogenases: in search of the active site. Biochim Biophys Acta. 1994;1188:167–204. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases