Characterization of a Bacteroides mobilizable transposon, NBU2, which carries a functional lincomycin resistance gene - PubMed (original) (raw)
Characterization of a Bacteroides mobilizable transposon, NBU2, which carries a functional lincomycin resistance gene
J Wang et al. J Bacteriol. 2000 Jun.
Abstract
The mobilizable Bacteroides element NBU2 (11 kbp) was found originally in two Bacteroides clinical isolates, Bacteroides fragilis ERL and B. thetaiotaomicron DOT. At first, NBU2 appeared to be very similar to another mobilizable Bacteroides element, NBU1, in a 2.5-kbp internal region, but further examination of the full DNA sequence of NBU2 now reveals that the region of near identity between NBU1 and NBU2 is limited to this small region and that, outside this region, there is little sequence similarity between the two elements. The integrase gene of NBU2, intN2, was located at one end of the element. This gene was necessary and sufficient for the integration of NBU2. The integrase of NBU2 has the conserved amino acids (R-H-R-Y) in the C-terminal end that are found in members of the lambda family of site-specific integrases. This was also the only region in which the NBU1 and NBU2 integrases shared any similarity (28% amino acid sequence identity and 49% sequence similarity). Integration of NBU2 was site specific in Bacteroides species. Integration occurred in two primary sites in B. thetaiotaomicron. Both of these sites were located in the 3' end of a serine-tRNA gene NBU2 also integrated in Escherichia coli, but integration was much less site specific than in B. thetaiotaomicron. Analysis of the sequence of NBU2 revealed two potential antibiotic resistance genes. The amino acid sequences of the putative proteins encoded by these genes had similarity to resistances found in gram-positive bacteria. Only one of these genes was expressed in B. thetaiotaomicron, the homolog of linA, a lincomycin resistance gene from Staphylococcus aureus. To determine how widespread elements related to NBU1 and NBU2 are in Bacteroides species, we screened 291 Bacteroides strains. Elements with some sequence similarity to NBU2 and NBU1 were widespread in Bacteroides strains, and the presence of linA(N) in Bacteroides strains was highly correlated with the presence of NBU2, suggesting that NBU2 has been responsible for the spread of this gene among Bacteroides strains. Our results suggest that the NBU-related elements form a large and heterogeneous family, whose members have similar integration mechanisms but have different target sites and differ in whether they carry resistance genes.
Figures
FIG. 1
Construction of the integration vector, pEPIntN2. An insertional vector that could be used in either Bacteroides or in E. coli was constructed from the pir_-dependent mobilizable pEP185.2 (24). The erythromycin-clindamycin resistance gene, ermG, from CTn_7853 was PCR amplified with _Pst_I sites encoded in the primers and cloned into the _Pst_I-compatible unique _Nsi_I site on pEP185.2 to form pEPE. pEPE, a Pir-dependent vector, has selectable markers for both E. coli and Bacteroides hosts. The NBU2 integration region, IntN2, consisting of the joined ends, attN2, from the circular form of NBU2 and the adjacent gene, intN2, was PCR amplified from the induced circular form of NBU2. The PCR product was first cloned into pGEM-T (Promega) and sequenced. The 1.8-kbp _Apa_I-_Sst_II fragment was isolated from pGEM-T and cloned into the _Apa_I and _Sst_II sites of pEPE to form the NBU2 integration vector, pEPIntN2.
FIG. 2
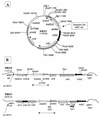
Circular and integrated forms of NBU2. (A) Partial restriction map of the excised circular form of NBU2. The location and orientation of the possible ORFs derived from the NBU2 sequence are indicated. The region containing the joined ends of the NBU2 (attN2) is indicated. The attN2 is contained within a 1.2-kbp _Hin_cII fragment. (B) The integrated form of NBU2 is compared to the integrated form of NBU1. The double-headed arrows indicate the region of high sequence identity between NBU2 and NBU1 (see Fig. 3). The attN-left sequences of the integrated NBUs are the same sequence as the attN on the elements, and the attN-right sequences are the attBT sequences of the target sites. Both elements integrate site specifically into the 3′ end of tRNA genes: Ser-tRNAUGA for NBU2 and Leu-tRNACAA for NBU1.
FIG. 3
Sequence comparison of NBU2 and NBU1. The prmN-oriT-mobN regions of NBU1 and NBU2 have more than 85% identity. Outside of this mobilization region the sequence identity drops to <30%. Near the borders of the sequence identities both elements have inverted repeat sequences (underlined) of 11 to 12 bp indicated by arrows above the sequences for NBU1 (IR1) and dotted arrows below the sequences for NBU2 (IR2). The start and stop codons for the prmN (PrmN) and mobN (MobN) are indicated in boldface. The oriT nick sites as determined by sequence identity to MTn_4555_ (53) are at the end of the TAG codon (Ω Stop) of the prmN genes and are indicated by the arrows.
FIG. 4
Southern blot analysis of the NBU2 insertions in Bacteroides attB2 sites. (A) Southern blot of two B. thetaiotaomicron BT4001 transconjugants from BT ERL that received both CTnERL and NBU2. The DNAs of the two strains were digested with _Hin_cII, and Southern blots were probed with the labeled 1.2-kbp _Hin_cII fragment containing attN2 (Fig. 2). BT4004N3 in lane 1 has a single insertion (two junction bands), and BT4004N6 in lane 2 has two insertions of NBU2 (four junction bands). In lane 3, BT4004N6 was grown in tetracycline to induce the circular excised form of NBU2. The 1.2-kbp fragment from the excised circular form of NBU2 is indicated by the arrow. (B) Southern blot showing 6 of 10 independent insertions of the minielement vector, pEPIntN2, into the B. thetaiotaomicron BT4001 chromosome. The DNAs from the strains were digested with _Apa_I-_Sst_II, and the Southern blot was probed with the labeled 1.8-kbp _Apa_I-_Sst_II fragment containing the _intN2_-attN2 region of NBU2 cloned on pEPIntN2 (Fig. 1). The sequence obtained for the cloned targets sites (see Materials and Methods) indicated that the insertion site of pEPIntN2 in lanes 1, 2, and 4 were the same as the wild-type element in BT4004N3 (panel A, lane 1). In BT4004N6, one of the ΩNBU2 copies was integrated into this site and the other copy of ΩNBU2 was integrated into the site represented by lanes 3, 5, and 6 of panel B. The locations of the _Hin_dIII lambda size fragments in kilobase pairs are marked on the left of each panel.
FIG. 5
C-terminal alignment of IntN2 and related members in the lambda family of site-specific integrases. The similarity of IntN2 to members of the lambda family of site-specific integrases was in the C-terminal end. The alignment of IntN2 and the integrases with the highest similarities is shown. Domain I and domain II in the C-terminal ends of this family of integrases contain conserved amino acids for the active sites of the integrases (2, 27). The conserved amino acids are labeled (#) and boxed for these active site domains. The total size of each integrase is shown at the end of its sequence as COOH-, with the number of amino acids in parentheses. The accession numbers for the integrases shown here are as follows: Bacteroides MTns NBU2 (L42370), NBU1 (L13840), and MTn_5520_ (AF038866) and cyanobacterial plasmid pDU1 (L23221), A. aeolicus plasmid, ece1, (C55205), L. lactis CTn_5276_ (L27649), bacteriophage P21 (P27077), and prophage E14 (M61865).
FIG. 6
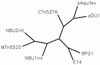
Unrooted tree of the NBU2 integrase and related integrases. The entire sequences of the lambda family of site-specific integrases, including the IntN2 of NBU2, that were aligned in Fig. 5 were grouped on a cladogram or unrooted tree to show their relationships using the Bioinformatics analyses conducted on BioNavigator.com provided by eBioinformatics. The tree showed three clusters: the three Bacteroides MTn integrases [NBU1 (IntN1), NBU2 (IntN2) and MTn_5520_ (IntBIP)], the two bacteriophage integrases from P21 and prophage E14, and the group containing the putative integrase-recombinases on plasmids from Nostoc (pDU1) and A. aeolicus plasmid ece1 (pAquifex) and the integrase from the L. lactis conjugative transposon CTn_5276_. Accession numbers are given in Fig. 5.
FIG. 7
NBU2 attN2 and chromosomal target sites. The attN2 region of NBU2, shown at the top of the figure, has two regions that have identity or similarity to its target sites. The crossovers occurred within or adjacent to a 13-bp sequence in highlighted capital letters that has sequence identity to the two attBT2 Bacteroides target sites. The second region of attN2 has a 14-bp sequence, boxed and highlighted, that has partial sequence identity to the attBT2 sites, and is located at the 5′ end of a pair of IRs. The IRs are indicated by arrows and the bases which could pair in a stem and loop structure are capitalized. The two attBT2 sites are located at the 3′ ends of Ser-tRNAUGA genes that differ from each other by only a single base pair. The crossovers occurred within or adjacent to the 13-bp sequence of identity to attN2. Both attBT2-1 and attBT2-2 also had 15-bp sequences with partial identity, boxed and highlighted, to the 14-bp sequence of attN2 that were located 5′ to IRs. The sequence for one of the pEPintN2 insertion sites in E. coli (attBEc) is shown at the bottom. The insertion occurred in the 3′ end of fecI. The crossover occurred adjacent to or within the triplet (CCT) at the beginning of a sequence with partial identity (8 of 13) to the 13-bp region on attN2. The crossover region for attBEc was followed by a set of IRs indicated by arrows which had no sequence identity to the NBU2 IR region.
FIG. 8
Alignment of the NBU2 lincosamide resistance, LinAN2, with LinA′ and LinA from Staphylococcus spp. The amino acid sequence of the lincosamide resistance on NBU2, LinA(N2), was aligned with the sequences of LinA′ from S. aureus and LinA from pIP855 in S. haemolyticus (5, 6) using the BLASTP search program. LinA(N2) has 52% identity and 70 to 72% identity to LinA′ and LinA. The regions of identity are indicated in boldface. The total number of amino acids for each protein is indicated at the ends of the respective sequences. The accession numbers for LinA′ and LinA are J03947 and A25633, respectively.
References
- Brisson-Noel A, Pl Delrieu, Samain D, Courvalin P. Inactivation of lincosaminide antibiotics in Staphylococcus. Identification of lincosaminide O-nucleotidyltransferases and comparison of the corresponding resistance genes. J Biol Chem. 1988;263:15880–15887. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical