A phosphoserine-regulated docking site in the protein kinase RSK2 that recruits and activates PDK1 - PubMed (original) (raw)
A phosphoserine-regulated docking site in the protein kinase RSK2 that recruits and activates PDK1
M Frödin et al. EMBO J. 2000.
Abstract
The 90 kDa ribosomal S6 kinase-2 (RSK2) is a growth factor-stimulated protein kinase with two kinase domains. The C-terminal kinase of RSK2 is activated by ERK-type MAP kinases, leading to autophosphorylation of RSK2 at Ser386 in a hydrophobic motif. The N-terminal kinase is activated by 3-phosphoinositide-dependent protein kinase-1 (PDK1) through phosphorylation of Ser227, and phosphorylates the substrates of RSK. Here, we identify Ser386 in the hydrophobic motif of RSK2 as a phosphorylation-dependent docking site and activator of PDK1. Treatment of cells with growth factor induced recruitment of PDK1 to the Ser386-phosphorylated hydrophobic motif and phosphorylation of RSK2 at Ser227. A RSK2-S386K mutant showed no interaction with PDK1 or phosphorylation at Ser227. Interaction with Ser386-phosphorylated RSK2 induced autophosphorylation of PDK1. Addition of a synthetic phosphoSer386 peptide (RSK2(373-396)) increased PDK1 activity 6-fold in vitro. Finally, mutants of RSK2 and MSK1, a RSK-related kinase, with increased affinity for PDK1, were constitutively active in vivo and phosphorylated histone H3. Our results suggest a novel regulatory mechanism based on phosphoserine-mediated recruitment of PDK1 to RSK2, leading to coordinated phosphorylation and activation of PDK1 and RSK2.
Figures
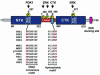
Fig. 1. Structure and regulatory phosphorylation sites of RSK2. RSK is composed of two kinase domains connected by a regulatory linker region. The C-terminal tail contains a docking site for ERK (Gavin and Nebreda, 1999; Smith et al., 1999). The locations of five phosphorylation sites and the kinases that phosphorylate these sites are shown. Phosphorylation at Ser227, Ser369, Ser386 and Thr577 regulates kinase activity, whereas the role of Thr365 phosphorylation is unclear (mouse RSK2 numbering). Amino acid sequences show the PDK1 consensus phosphorylation motif and the hydrophobic motif with conserved residues in bold. The alignment illustrates that both motifs are present in many growth factor-activated kinases, including RSK, MSK, p70 S6K, PKBα, PKCδ, SGK and PRK2. Serines/threonines that require phosphorylation for kinase activation are shown in red. Sequences are human, except RSK1 (rat), RSK2 and PKCδ (both mouse).
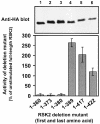
Fig. 2. Kinase activity of RSK2 deletion mutants in vivo. COS7 cells were transfected with plasmids expressing HA epitope-tagged full-length RSK2 or deletion mutants. After 48 h and a final 3 h serum starvation period, cells were lysed and RSK was precipitated with Ab to the HA epitope tag and subjected to kinase assay (lower panel). Data are expressed as a percentage of full-length RSK2 from unstimulated cells and are the mean ± SD of three independent experiments performed in duplicate. The activities of RSK21–389 and unstimulated RSK2 were different (p<0.001) when compared by non-paired _t_-test. After the kinase assay, RSK was subjected to SDS–PAGE and immunoblotting with Ab to the HA epitope tag (upper panel).
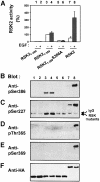
Fig. 3. Role of the hydrophobic motif in regulation of phosphorylation sites in the N-terminal kinase of RSK2. COS7 cells were transfected with plasmids expressing HA epitope-tagged wild-type RSK2 or mutant RSK2. After 48 h and a final 3 h serum starvation period, cells were exposed, or not, to 20 nM EGF for 20 min and lysed. Thereafter, RSK was precipitated from the cell lysates with Ab to the HA epitope. (A) The kinase activity of RSK was determined and expressed as a percentage of basal RSK21–389 activity. Data are the mean ± SD of four independent experiments performed in duplicate. The data of the following bars were different when compared by non-paired _t_-test: 3 versus 5 (p <0.001); 3 versus 7 (p <0.05). After the kinase assay, aliquots of the precipitated RSKs were subjected to SDS–PAGE and immunoblotting with Abs that bind RSK2 when phosphorylated at Ser386 (B), Ser227 (C), Thr365 (D) or Ser369 (E), or with Ab to the HA tag (F).
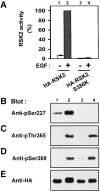
Fig. 4. Ser386 is required for phosphorylation of Ser227 in RSK2. COS7 cells were transfected with plasmids expressing HA-tagged RSK2 or RSK2-S386K. After 48 h and a final 3 h serum starvation period, cells were exposed, or not, to 20 nM EGF for 20 min and lysed. (A) RSK was precipitated from the cell lysates with Ab to the HA tag, its kinase activity was determined and expressed as a percentage of EGF-stimulated wild-type RSK2 activity. Data are the mean ± SD of three independent experiments performed in duplicate. After the kinase assay, aliquots of the precipitated RSKs were subjected to SDS–PAGE and immunoblotting with Abs that bind RSK2 when phosphorylated at Ser227 (B), Thr365 (C), Ser369 (D) or with Ab to the HA epitope tag (E).
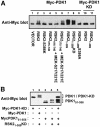
Fig. 5. Ser386 is required for RSK2 to interact with PDK1. COS7 cells were co-transfected with plasmids expressing myc-PDK1 and HA-RSK2 or HA-RSK2-S386K. After 48 h and a final 3 h serum starvation period, the cells were lysed and cell lysates were subjected to immunoprecipitation with Ab to the HA tag. Aliquots of the precipitates were subjected to SDS–PAGE and immunoblotting with Ab to the myc epitope tag on PDK1 (A) or to the HA epitope tag on RSK2 (B). Equal expression of PDK1 in the cells was verified by subjecting pre-immunoprecipitation lysates to immunoblotting for the myc tag (C). The experiments were performed three times with similar results.
Fig. 6. PDK1 interacts with RSK2 via the Ser386-phosphorylated hydrophobic motif. COS7 cells were transfected with plasmids expressing myc-PDK1, HA-RSK2, HA-RSK2 mutant, MEK-S217/221E or with empty vector (–), as indicated in each panel. After 48 h and a final 3 h serum starvation period, the cells were lysed. (A) Cell lysates were subjected to immunoprecipitation with Ab to the HA tag, whereafter aliquots of the precipitates were subjected to SDS–PAGE and immunoblotting with Ab to the myc tag in PDK1 or to the HA tag in RSK2. (B) Cell lysates were subjected to immunoprecipitation with Ab to the HA tag in lysis buffer with or without phosphatase inhibitors (orthovanadate, calyculin, sodium flouride). Aliquots of the precipitates were subjected to SDS–PAGE and immunoblotting with Ab to the myc tag in PDK1, to phosphoSer386 in RSK2 or to the HA tag in RSK2. (C) Cell lysates were subjected to immunoprecipitation with Ab to the CTK domain of RSK2. Aliquots of the precipitates were subjected to SDS–PAGE and immunoblotting with Ab to the myc tag in PDK1, to phosphoSer386 in RSK2 or to the HA tag in RSK2. The experiments were performed three times with similar results.
Fig. 6. PDK1 interacts with RSK2 via the Ser386-phosphorylated hydrophobic motif. COS7 cells were transfected with plasmids expressing myc-PDK1, HA-RSK2, HA-RSK2 mutant, MEK-S217/221E or with empty vector (–), as indicated in each panel. After 48 h and a final 3 h serum starvation period, the cells were lysed. (A) Cell lysates were subjected to immunoprecipitation with Ab to the HA tag, whereafter aliquots of the precipitates were subjected to SDS–PAGE and immunoblotting with Ab to the myc tag in PDK1 or to the HA tag in RSK2. (B) Cell lysates were subjected to immunoprecipitation with Ab to the HA tag in lysis buffer with or without phosphatase inhibitors (orthovanadate, calyculin, sodium flouride). Aliquots of the precipitates were subjected to SDS–PAGE and immunoblotting with Ab to the myc tag in PDK1, to phosphoSer386 in RSK2 or to the HA tag in RSK2. (C) Cell lysates were subjected to immunoprecipitation with Ab to the CTK domain of RSK2. Aliquots of the precipitates were subjected to SDS–PAGE and immunoblotting with Ab to the myc tag in PDK1, to phosphoSer386 in RSK2 or to the HA tag in RSK2. The experiments were performed three times with similar results.
Fig. 7. Recruitment of PDK1 to RSK2 in growth factor-treated cells. COS7 cells were co-transfected with plasmids expressing kinase-deficient (KD) myc-PDK1 and HA-RSK2 or HA-RSK2-S386K, as indicated. After 48 h and a final 3 h serum starvation period, cells were exposed, or not, to 20 nM EGF for 25 min and then lysed. The cell lysates were subjected to immunoprecipitation with Ab to the HA tag. Aliquots of the precipitates were subjected to SDS–PAGE and immunoblotting with Ab to the myc tag in PDK1 (A) or to the HA tag in RSK2 (B). Equal expression of PDK1 in the cells was verified by subjecting pre-immunoprecipitation lysates to immunoblotting for the myc tag (C). The experiments were performed three times with similar results.
Fig. 8. Autophosphorylation of PDK1 after binding to the Ser386-phosphorylated hydrophobic motif of RSK2 in vivo. (A and B) COS7 cells were co-transfected with plasmids expressing wild-type or mutant forms of myc-PDK1 and HA-RSK2 and with MEK-S217/221E as indicated. KD denotes kinase-deficient mutant. Forty-eight hours after transfection, cells were serum starved for 3 h. Thereafter, cells were lysed in SDS–PAGE sample buffer and subjected to SDS–PAGE and immunoblotting with Ab to the myc tag in PDK1. The experiments were performed three times with similar results.
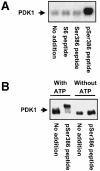
Fig. 9. Autophosphorylation of PDK1 induced by an RSK2 peptide containing phospho-Ser386 in vitro. Immunopurified myc-PDK1 was pre-incubated for 20 min alone (no addition) or with 10 µM S6 peptide or RSK2 peptide, residues 373–396, which was unphosphorylated (Ser386 peptide) or phosphorylated at Ser386 (pSer386 peptide). (A) PDK1 was allowed to autophosphorylate for 20 min in the presence of [γ-32P]ATP, whereafter the reactions were subjected to SDS–PAGE and autoradiography. (B) PDK1 was allowed to autophosphorylate for 35 min in the presence or absence of ATP, whereafter the reactions were subjected to SDS–PAGE and immunoblotting with Ab to the myc tag in PDK1. The experiments were performed twice with similar results.
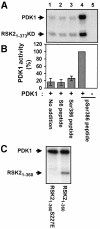
Fig. 10. Activation of PDK1 by a RSK2 peptide containing phosphoSer386 in vitro. Immunopurified myc-PDK1 was pre-incubated for 20 min alone (no addition) or with 10 µM S6 peptide, Ser386 peptide or pSer386 peptide. (A and C) PDK1 was then incubated for 10 min with [γ-32P]ATP and 2 µg of RSK21–373KD or 0.5 µg of RSK21–360 as a substrate, as indicated in each panel, whereafter the reactions were subjected to SDS–PAGE and autoradiography. (B) PDK1 activity in (A) was determined by quantitation of radioactivity incorporated into RSK21–373KD using a PhosphorImager. Data are expressed as a percentage of PDK1 activity in the presence of pSer386 peptide and are the mean ± SD of three independent experiments. The data of the following bars were not different when compared by non-paired _t_-test: 1 versus 2 or 3 (p >0.2). The data from bars 1 and 4 were different when compared by non-paired _t_-test (p <0.001).
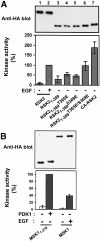
Fig. 11. Constitutively active mutants of RSK2 and MSK1. COS7 cells were transfected with plasmids expressing HA-tagged wild-type or mutant RSK2 or MSK1 and myc-tagged PDK1 as indicated in each panel. After 48 h and a final 3 h serum starvation period, cells were exposed, or not, to 20 nM EGF for 20 min and lysed. (A) RSK2 was precipitated from the cell lysates with Ab to the HA tag and subjected to kinase assay (lower panel). Data are expressed as a percentage of EGF-stimulated full-length RSK2 activity and are the mean ± SD of 3–4 independent experiments performed in duplicate The data of the following bars were different when compared by non-paired _t_-test: 3 versus 6 (p <0.05); 3 versus 7 (p <0.01). After the kinase assay, the precipitates were subjected to immunoblotting with Ab to the HA tag (upper panel). (B) MSK1 was precipitated from cell lysates with Ab to the HA tag, its kinase activity was determined and expressed as a percentage of EGF-stimulated wild-type MSK1 activity (lower panel). Data are the mean ± SD of four independent experiments performed in duplicate. After the kinase assay, MSK was subjected to immunoblotting with Ab to the HA tag (upper panel). (C) MSK1 mutants were precipitated from the cell lysates with Ab to the HA tag. Precipitates were subjected to immunoblotting with Ab to the myc tag in PDK1 or to the HA tag in MSK1. Equal expression of PDK1 in the cells was verified by subjecting pre-immunoprecipitation lysates to immunoblotting for the myc tag. The experiment was performed three times with similar results. (D) MSK1 mutants were precipitated from cell lysates with Ab to the HA tag and subjected to kinase assay (lower panel). Data are expressed as a percentage of CA-MSK1 activity and are the mean ± SD of three independent experiments performed in duplicate. After the kinase assay, the precipitates were subjected to immunoblotting with Ab to the HA tag (upper panel).
Fig. 11. Constitutively active mutants of RSK2 and MSK1. COS7 cells were transfected with plasmids expressing HA-tagged wild-type or mutant RSK2 or MSK1 and myc-tagged PDK1 as indicated in each panel. After 48 h and a final 3 h serum starvation period, cells were exposed, or not, to 20 nM EGF for 20 min and lysed. (A) RSK2 was precipitated from the cell lysates with Ab to the HA tag and subjected to kinase assay (lower panel). Data are expressed as a percentage of EGF-stimulated full-length RSK2 activity and are the mean ± SD of 3–4 independent experiments performed in duplicate The data of the following bars were different when compared by non-paired _t_-test: 3 versus 6 (p <0.05); 3 versus 7 (p <0.01). After the kinase assay, the precipitates were subjected to immunoblotting with Ab to the HA tag (upper panel). (B) MSK1 was precipitated from cell lysates with Ab to the HA tag, its kinase activity was determined and expressed as a percentage of EGF-stimulated wild-type MSK1 activity (lower panel). Data are the mean ± SD of four independent experiments performed in duplicate. After the kinase assay, MSK was subjected to immunoblotting with Ab to the HA tag (upper panel). (C) MSK1 mutants were precipitated from the cell lysates with Ab to the HA tag. Precipitates were subjected to immunoblotting with Ab to the myc tag in PDK1 or to the HA tag in MSK1. Equal expression of PDK1 in the cells was verified by subjecting pre-immunoprecipitation lysates to immunoblotting for the myc tag. The experiment was performed three times with similar results. (D) MSK1 mutants were precipitated from cell lysates with Ab to the HA tag and subjected to kinase assay (lower panel). Data are expressed as a percentage of CA-MSK1 activity and are the mean ± SD of three independent experiments performed in duplicate. After the kinase assay, the precipitates were subjected to immunoblotting with Ab to the HA tag (upper panel).
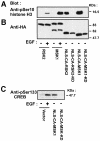
Fig. 12. In vivo phosphorylation of histone H3 and CREB by nuclear-targeted, constitutively active mutants of RSK2 and MSK1. COS7 cells were transfected with plasmids expressing HA-tagged wild-type or mutant RSK2 or MSK1. NLS, nuclear localization sequence. After 24 h and a final 3 h serum starvation period, cells were exposed, or not, to 20 nM EGF for 25 min. Cells were lysed in SDS–PAGE sample buffer and subjected to immunoblotting with Abs to histone H3 phosphorylated at Ser10 (A), to the HA tag (B) or to CREB phosphorylated at Ser133 (C).
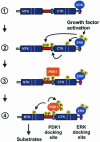
Fig. 13. Model for recruitment and coordinated activation of PDK1 and RSK2. (1) Quiescent cells contain a pre-formed complex of inactive RSK and ERK. (2) Stimulation of the Ras–ERK pathway results in ERK-catalyzed phosphorylation of the linker and the CTK, leading to autophosphorylation of RSK2 at Ser386 in the hydrophobic motif (blue box). (3) Phosphorylation of Ser386 in the hydrophobic motif creates a docking site that allows complex formation between PDK1 and RSK2. (4) Interaction of PDK1 with the Ser386-phosphorylated hydrophobic motif stimulates PDK1 activity and autophosphorylation. The combination of increased local concentration and kinase activity of PDK1 ensures efficient phosphorylation of Ser227, leading to full activation of RSK.
Similar articles
- p90 ribosomal S6 kinase 2 is associated with and dephosphorylated by protein phosphatase 2Cdelta.
Doehn U, Gammeltoft S, Shen SH, Jensen CJ. Doehn U, et al. Biochem J. 2004 Sep 1;382(Pt 2):425-31. doi: 10.1042/BJ20040948. Biochem J. 2004. PMID: 15206906 Free PMC article. - 90-kDa ribosomal S6 kinase is phosphorylated and activated by 3-phosphoinositide-dependent protein kinase-1.
Jensen CJ, Buch MB, Krag TO, Hemmings BA, Gammeltoft S, Frödin M. Jensen CJ, et al. J Biol Chem. 1999 Sep 17;274(38):27168-76. doi: 10.1074/jbc.274.38.27168. J Biol Chem. 1999. PMID: 10480933 - A phosphoserine/threonine-binding pocket in AGC kinases and PDK1 mediates activation by hydrophobic motif phosphorylation.
Frödin M, Antal TL, Dümmler BA, Jensen CJ, Deak M, Gammeltoft S, Biondi RM. Frödin M, et al. EMBO J. 2002 Oct 15;21(20):5396-407. doi: 10.1093/emboj/cdf551. EMBO J. 2002. PMID: 12374740 Free PMC article. - Signalling specificity of Ser/Thr protein kinases through docking-site-mediated interactions.
Biondi RM, Nebreda AR. Biondi RM, et al. Biochem J. 2003 May 15;372(Pt 1):1-13. doi: 10.1042/BJ20021641. Biochem J. 2003. PMID: 12600273 Free PMC article. Review. - Targeting RSK2 in Cancer Therapy: A Review of Natural Products.
Wu T, Chen Z, Liu X, Wu X, Wang Z, Guo W. Wu T, et al. Anticancer Agents Med Chem. 2025;25(1):35-41. doi: 10.2174/0118715206329546240830055233. Anticancer Agents Med Chem. 2025. PMID: 39248063 Review.
Cited by
- Essential role of PDK1 in regulating cell size and development in mice.
Lawlor MA, Mora A, Ashby PR, Williams MR, Murray-Tait V, Malone L, Prescott AR, Lucocq JM, Alessi DR. Lawlor MA, et al. EMBO J. 2002 Jul 15;21(14):3728-38. doi: 10.1093/emboj/cdf387. EMBO J. 2002. PMID: 12110585 Free PMC article. - RNA interference screening identifies lenalidomide sensitizers in multiple myeloma, including RSK2.
Zhu YX, Yin H, Bruins LA, Shi CX, Jedlowski P, Aziz M, Sereduk C, Kortuem KM, Schmidt JE, Champion M, Braggio E, Keith Stewart A. Zhu YX, et al. Blood. 2015 Jan 15;125(3):483-91. doi: 10.1182/blood-2014-05-577130. Epub 2014 Nov 13. Blood. 2015. PMID: 25395420 Free PMC article. - RSK2 activity mediates glioblastoma invasiveness and is a potential target for new therapeutics.
Sulzmaier FJ, Young-Robbins S, Jiang P, Geerts D, Prechtl AM, Matter ML, Kesari S, Ramos JW. Sulzmaier FJ, et al. Oncotarget. 2016 Nov 29;7(48):79869-79884. doi: 10.18632/oncotarget.13084. Oncotarget. 2016. PMID: 27829215 Free PMC article. - RSK Isoforms in Acute Myeloid Leukemia.
Youn M, Gomez JO, Mark K, Sakamoto KM. Youn M, et al. Biomedicines. 2021 Jun 24;9(7):726. doi: 10.3390/biomedicines9070726. Biomedicines. 2021. PMID: 34202904 Free PMC article. Review. - Protein phosphatase 2Cδ/Wip1 regulates phospho-p90RSK2 activity in lesional psoriatic skin.
Rasmussen MK, Nielsen J, Kjellerup RB, Andersen SM, Rittig AH, Johansen C, Iversen L, Gesser B. Rasmussen MK, et al. J Inflamm Res. 2017 Dec 15;10:169-180. doi: 10.2147/JIR.S152869. eCollection 2017. J Inflamm Res. 2017. PMID: 29290690 Free PMC article.
References
- Alessi D.R. et al. (1997a) 3-Phosphoinositide-dependent protein kinase-1 (PDK1): structural and functional homology with the Drosophila DSTPK61 kinase. Curr. Biol., 7, 776–789. - PubMed
- Alessi D.R., Kozlowski,M.T., Weng,Q.-P., Morrice,N. and Avruch,J. (1997b) 3-Phosphoinositide-dependent protein kinase 1 (PDK1) phosphorylates and activates the p70 S6 kinase in vivo and in vitro.Curr. Biol., 8, 69–81. - PubMed
- Anderson K.E., Coadwell,J., Stephens,L.R. and Hawkins,P.T. (1998) Translocation of PDK-1 to the plasma membrane is important in allowing PDK-1 to activate protein kinase B. Curr. Biol., 8, 684–691. - PubMed
- Andjelkovic M. et al. (1997) Role of translocation in the activation and function of protein kinase B. J. Biol. Chem., 272, 31515–31524. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous