Overlapping roles for the histone acetyltransferase activities of SAGA and elongator in vivo - PubMed (original) (raw)
Overlapping roles for the histone acetyltransferase activities of SAGA and elongator in vivo
B O Wittschieben et al. EMBO J. 2000.
Abstract
Elp3 and Gcn5 are histone acetyltransferases (HATs) that function in transcription as subunits of Elongator and SAGA/ADA, respectively. Here we show that mutations that impair the in vitro HAT activity of Elp3 confer typical elp phenotypes such as temperature sensitivity. Combining an elp3Delta mutation with histone H3 or H4 tail mutations confers lethality or sickness, supporting a role for Elongator in chromatin remodelling in vivo. gcn5Deltaelp3Delta double mutants display a number of severe phenotypes, and similar phenotypes result from combining the elp mutation with mutation in a gene encoding a SAGA-specific, but not an ADA-specific subunit, indicating that Elongator functionally overlaps with SAGA. Because concomitant active site alterations in Elp3 and Gcn5 are sufficient to confer severe phenotypes, the redundancy must be specifically related to the HAT activity of these complexes. In support of this conclusion, gcn5Deltaelp3Delta phenotypes are suppressed by concomitant mutation of the HDA1 and HOS2 histone deacetylases. Our results demonstrate functional redundancy among transcription-associated HAT and deacetylase activities, and indicate the importance of a fine-tuned acetylation-deacetylation balance during transcription in vivo.
Figures
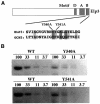
Fig. 1. Elp3 mutants have reduced HAT activity in vitro. (A) Alignment of Elp3 and Gcn5 HAT motif B based on Neuwald and Landsman (1997) with the positions of the changes introduced in Elp3 indicated. (B) The HAT activity of Elp3 mutants is significantly reduced. Titrations of equal amounts of wild-type and mutant proteins were fractionated in 10% SDS–PAGE gels containing either BSA (not shown) or 1 mg/ml histone. After renaturation of separated proteins, the gels were incubated with tritium-labeled acetyl-CoA. Autoradiograms of dried gels are shown. Relative amounts of protein loaded are indicated above the lanes. Based on two independent titration experiments, the Y540A mutant had 22–25% activity, while Y541A had 34–38% activity compared with wild type.
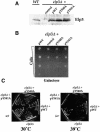
Fig. 2. Elp3 HAT mutants display elp phenotypes. (A) Similar levels of wild-type and mutant Elp3 protein are detected in whole-cells extracts. Extracts from elp3Δ cells carrying CEN plasmids expressing wild-type Elp3 (pWT), no protein (control), Elp3 Y540A (pY540A) or Elp3 Y541A (pY541A) were compared with wild-type (ELP3) extracts. The result of a western blot probed with anti-Elp3 antibodies is shown. (B) Elp3 protein-carrying altered HAT domains are unable to complement the slow adaptation phenotype typical of elpΔ strains. Strains carrying the above CEN plasmids were grown in YPD and plated in a dilution series onto YPG (galactose) plates and incubated at 30°C for 2–3 days. (C) Elp3 protein-carrying altered HAT domains are unable to complement the temperature-sensitive phenotype of an elp3Δ strain. Strains were treated as in (B), but streaked on YPD plates and incubated for 3–5 days at 30 or 39°C.
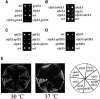
Fig. 3. Synthetic phenotype of the elp3Δgcn5Δ strain. (A) elp3Δgcn5Δ double mutants are slow growing. Tetrad dissections of gcn5Δ/GCN5 elp3Δ/ELP3 diploids are shown after 2 days of incubation at 30°C. (B) elp3Δgcn5Δ double mutants are unable to grow on a number of media. Cells of the indicated genotype were streaked on YPD [glucose (30°C)], YPG (galactose), YPS (sucrose) or inositol starvation (no inositol) plates, and incubated for 3–5 days at 30°C. To demonstrate temperature sensitivity, cells of the indicated genotype were streaked on YPD [37°C (glucose)] and incubated for 3–5 days at 37°C.
Fig. 4. Severe phenotypes of cells with concomitant disruptions of Elongator and SAGA genes. (A–D) elp1Δgcn5Δ and elp3Δspt20Δ, but not elp3Δahc1Δ and elp3Δspt3Δ double mutants are slow growing. Tetrad dissections of the respective diploids are shown after 2–3 days of incubation at 30°C. (E) elp3Δspt3Δ, but not elp1Δgcn5Δ, elp2Δgcn5Δ and elp3Δspt20Δ double mutants grow at 37°C. Cells of the indicated genotype were streaked on YPD and incubated for 3–5 days at 30 or 37°C. As expected, elp3Δahc1Δ cells are not temperature-sensitive either (data not shown).
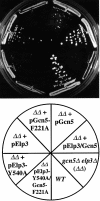
Fig. 5. Severe phenotypes of cells with concomitant disruption of Elp3 and Gcn5 HAT activity. The effects of altered HAT domains in GCN5 and ELP3 were tested in gcn5Δelp3Δ cells carrying wild-type ELP3 on a _URA3_-marked plasmid. Cells capable of losing the URA3 plasmid were grown on 5-FOA-containing synthetic media for 4–6 days at 30°C. See Materials and methods for details. The slight growth of gcn5Δelp3Δ cells carrying pELP3-Y540A was not observed in other similar experiments. gcn5Δelp3Δ cells expressing altered versions of the HAT domains were also temperature-sensitive and unable to grow on alternative carbon sources (data not shown).
Fig. 6. The severe consequences of the gcn5Δelp3Δ mutation can be suppressed by mutation of specific HDACs. Mutation of HDA1 and HDA1 HOS2 suppresses the severe consequences of the gcn5Δelp3Δ mutation. Cells of the indicated genotype were streaked on YPG or YPS and incubated for 3–4 days at 30°C. Mutation of HDA1 and HOS2 also suppresses the temperature sensitivity (37°C) as well as the slow growth on YPD (30°C) of gcn5Δelp3Δ cells (data not shown).
Similar articles
- Histone H3 specific acetyltransferases are essential for cell cycle progression.
Howe L, Auston D, Grant P, John S, Cook RG, Workman JL, Pillus L. Howe L, et al. Genes Dev. 2001 Dec 1;15(23):3144-54. doi: 10.1101/gad.931401. Genes Dev. 2001. PMID: 11731478 Free PMC article. - Recruitment of chromatin remodelling factors during gene activation via the glucocorticoid receptor N-terminal domain.
Wallberg AE, Flinn EM, Gustafsson JA, Wright AP. Wallberg AE, et al. Biochem Soc Trans. 2000;28(4):410-4. Biochem Soc Trans. 2000. PMID: 10961930 Review. - Elongator is a histone H3 and H4 acetyltransferase important for normal histone acetylation levels in vivo.
Winkler GS, Kristjuhan A, Erdjument-Bromage H, Tempst P, Svejstrup JQ. Winkler GS, et al. Proc Natl Acad Sci U S A. 2002 Mar 19;99(6):3517-22. doi: 10.1073/pnas.022042899. Proc Natl Acad Sci U S A. 2002. PMID: 11904415 Free PMC article. - Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex.
Grant PA, Duggan L, Côté J, Roberts SM, Brownell JE, Candau R, Ohba R, Owen-Hughes T, Allis CD, Winston F, Berger SL, Workman JL. Grant PA, et al. Genes Dev. 1997 Jul 1;11(13):1640-50. doi: 10.1101/gad.11.13.1640. Genes Dev. 1997. PMID: 9224714 - A SAGA of histone acetylation and gene expression.
Hampsey M. Hampsey M. Trends Genet. 1997 Nov;13(11):427-9. doi: 10.1016/s0168-9525(97)01292-4. Trends Genet. 1997. PMID: 9385836 Review. No abstract available.
Cited by
- Functional Characterization of the GNAT Family Histone Acetyltransferase Elp3 and GcnE in Aspergillus fumigatus.
Choi YH, Park SH, Kim SS, Lee MW, Yu JH, Shin KS. Choi YH, et al. Int J Mol Sci. 2023 Jan 22;24(3):2179. doi: 10.3390/ijms24032179. Int J Mol Sci. 2023. PMID: 36768506 Free PMC article. - The Elongator Subunit Elp3 Regulates Development, Stress Tolerance, Cell Cycle, and Virulence in the Entomopathogenic Fungus Beauveria bassiana.
Cai Q, Wang J, Xie J, Jiang D, Keyhani NO. Cai Q, et al. J Fungi (Basel). 2022 Aug 10;8(8):834. doi: 10.3390/jof8080834. J Fungi (Basel). 2022. PMID: 36012822 Free PMC article. - Development of Artificial System to Induce Chromatin Loosening in Saccharomyces cerevisiae.
Yamamoto R, Sato G, Amai T, Ueda M, Kuroda K. Yamamoto R, et al. Biomolecules. 2022 Aug 18;12(8):1138. doi: 10.3390/biom12081138. Biomolecules. 2022. PMID: 36009033 Free PMC article. - Anticodon Wobble Uridine Modification by Elongator at the Crossroad of Cell Signaling, Differentiation, and Diseases.
Hermand D. Hermand D. Epigenomes. 2020 May 12;4(2):7. doi: 10.3390/epigenomes4020007. Epigenomes. 2020. PMID: 34968241 Free PMC article. Review. - The biogenesis and function of nucleosome arrays.
Singh AK, Schauer T, Pfaller L, Straub T, Mueller-Planitz F. Singh AK, et al. Nat Commun. 2021 Dec 1;12(1):7011. doi: 10.1038/s41467-021-27285-6. Nat Commun. 2021. PMID: 34853297 Free PMC article.
References
- Angus-Hill M.L., Dutnall,R.N., Tafrov,S.T., Sternglanz,R. and Ramakrishnan,V. (1999) Crystal structure of the histone acetyltransferase Hpa2: a tetrameric member of the Gcn5-related N-acetyltransferase superfamily. J. Mol. Biol., 294, 1311–1325. - PubMed
- Ayer D.E. (1999) Histone deacetylases: transcriptional repression with SINers and NuRDs. Trends Cell Biol., 9, 193–198. - PubMed
- Breeden L. and Nasmyth,K. (1987) Cell cycle control of the yeast HO gene: cis- and trans-acting regulators. Cell, 48, 389–397. - PubMed
- Brown C.E., Lechner,T., Howe,L. and Workman,J.L. (2000) The many HATs of transcription coactivators. Trends Biochem. Sci., 25, 15–19. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials