Inhibitory and bactericidal effects of hydrogen peroxide production by Streptococcus pneumoniae on other inhabitants of the upper respiratory tract - PubMed (original) (raw)
Inhibitory and bactericidal effects of hydrogen peroxide production by Streptococcus pneumoniae on other inhabitants of the upper respiratory tract
C D Pericone et al. Infect Immun. 2000 Jul.
Abstract
An inverse correlation between colonization of the human nasopharynx by Streptococcus pneumoniae and Haemophilus influenzae, both common upper respiratory pathogens, has been reported. Studies were undertaken to determine if either of these organisms produces substances which inhibit growth of the other. Culture supernatants from S. pneumoniae inhibited growth of H. influenzae, whereas culture supernatants from H. influenzae had no effect on the growth of S. pneumoniae. Moreover, coculture of S. pneumoniae and H. influenzae led to a rapid decrease in viable counts of H. influenzae. The addition of purified catalase prevented killing of H. influenzae in coculture experiments, suggesting that hydrogen peroxide may be responsible for this bactericidal activity. H. influenzae was killed by concentrations of hydrogen peroxide similar to that produced by S. pneumoniae. Hydrogen peroxide is produced by the pneumococcus through the action of pyruvate oxidase (SpxB) under conditions of aerobic growth. Both an spxB mutant and a naturally occurring variant of S. pneumoniae, which is downregulated in SpxB expression, were unable to kill H. influenzae. A catalase-reversible inhibitory effect of S. pneumoniae on the growth of the respiratory tract pathogens Moraxella catarrhalis and Neisseria meningitidis was also observed. Elevated hydrogen peroxide production, therefore, may be a means by which S. pneumoniae is able to inhibit a variety of competing organisms in the aerobic environment of the upper respiratory tract.
Figures
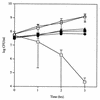
FIG. 1
Effect of coculture of S. pneumoniae P394 and H. influenzae Rd. Following growth to mid-log phase, H. influenzae was washed and incubated in sBHI containing heat-inactivated catalase either with (□) or without (○) S. pneumoniae for the time indicated, and viable counts were determined in duplicate on selective media. Viable counts of S. pneumoniae incubated in coculture with (■) or without (●) H. influenzae were determined in duplicate by plating on selective media. The same amount of active catalase (1,000 U/ml) was included during coculture of S. pneumoniae (▴) and H. influenzae (▵). Values represent the average of three independent determinations in duplicate, and the error bars represent the standard deviations.
FIG. 2
Dose-dependent killing of H. influenzae Rd by S. pneumoniae P394. Following growth to mid-log phase, H. influenzae was washed and cultured alone (triangles) or with 105 (squares), 106 (circles), or 107 (diamonds) CFU of S. pneumoniae per ml and incubated in sBHI for the times indicated; viable counts were determined on selective media. Values represent the average of two independent determinations in duplicate.
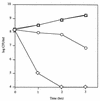
FIG. 3
Effect of coculture of H. influenzae Rd with S. pneumoniae D39 and its spxB mutant, P878. Following growth to mid-log phase, H. influenzae was washed and incubated in sBHI alone (○), with D39 (□), or with P878 (▵) for the times indicated, and viable counts were determined in duplicate on selective media. Viable counts of D39 (■) or P878 (▴) incubated in coculture with H. influenzae were determined in duplicate by plating on selective media. Values represent the average of three independent determinations in duplicate, and the error bars represent the standard deviations.
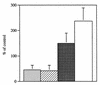
FIG. 4
Effect of coculture of S. pneumoniae P394 with either M. catarrhalis (Bc1) or N. meningitidis (MC58C3). Following growth to mid-log phase, S. pneumoniae (P394) was washed and incubated in BHI alone, with N. meningitidis (MC58C3) for 1.5 h, or with M. catarrhalis (Bc1) for 3 h. Viable counts of N. meningitidis (stippled bar) or M. catarrhalis (hatched bar) incubated in coculture with S. pneumoniae were determined in duplicate by plating on selective media. Viable counts of S. pneumoniae in coculture with N. meningitidis (black bar) or M. catarrhalis (white bar) were determined in duplicate on selective media. Values represent the change in viable count expressed as a percentage of a control culture containing that organism alone. Values are the average of three experiments, and error bars represent the standard deviations.
FIG. 5
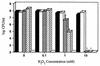
Effect of H2O2 on the survival of S. pneumoniae (P394), M. catarrhalis (Bc1), N. meningitidis (MC58C3), and H. influenzae (Rd). Following growth to mid-log phase, S. pneumoniae (black bars), M. catarrhalis (white bars), N. meningitidis (stippled bars), or H. influenzae (hatched bars) were washed and incubated at 37°C in BHI or sBHI containing the indicated concentration of H2O2. After 30 min, viable counts were determined on BHI or sBHI plates containing 200 U of catalase per ml. Values represent the average of three independent determinations in duplicate, and the error bars represent the standard deviations. ∗, Below the limit of detection.
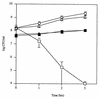
FIG. 6
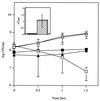
Effect of coculture of H. influenzae Rd with S. pneumoniae opaque (P62) or transparent (P64) variants of a type 9V isolate. Following growth to mid-log phase, H. influenzae was washed and incubated in sBHI either alone (○), with P62 (▵), or with P64 (□) for the times indicated, and viable counts were determined in duplicate on selective media. Viable counts of P62 (▴) or P64 (■) incubated in coculture with H. influenzae were determined in duplicate by plating on selective media. Values represent the average of three independent determinations in duplicate, and the error bars represent the standard deviations. (Inset) Relative expression of SpxB in S. pneumoniae variants P62 (black bar) and P64 (hatched bar) as determined by two-dimensional gel electrophoresis followed by mass spectrometric analysis. Results represent the average of four independent experiments, with error bars representing the standard deviations.
FIG. 7
Western blot showing the effect of environmental oxygen and carbon dioxide tension on pyruvate oxidase (SpxB) expression in S. pneumoniae P878, which contains an in-frame fusion to PhoA. Cell lysates of spxB::phoA mutant (P878) grown under 20% O2–0.03% CO2 (lane 1), 17% O2–3% CO2 (lane 2), or <0.01% O2–10% CO2 (lane 3) were electrophoresed on an SDS–10% polyacrylamide gel, transferred to a polyvinylidene difluoride membrane, and immunoblotted with an antibody to PhoA. As a negative control, cell lysates from the parent strain (D39) grown under 17% O2–3% CO2 (lane 4) were included. Size markers are in kilodaltons.
Similar articles
- Factors contributing to hydrogen peroxide resistance in Streptococcus pneumoniae include pyruvate oxidase (SpxB) and avoidance of the toxic effects of the fenton reaction.
Pericone CD, Park S, Imlay JA, Weiser JN. Pericone CD, et al. J Bacteriol. 2003 Dec;185(23):6815-25. doi: 10.1128/JB.185.23.6815-6825.2003. J Bacteriol. 2003. PMID: 14617646 Free PMC article. - Susceptibilities to levofloxacin in Streptococcus pneumoniae, Haemophilus influenzae, and Moraxella catarrhalis clinical isolates from children: results from 2000-2001 and 2001-2002 TRUST studies in the United States.
Karlowsky JA, Thornsberry C, Critchley IA, Jones ME, Evangelista AT, Noel GJ, Sahm DF. Karlowsky JA, et al. Antimicrob Agents Chemother. 2003 Jun;47(6):1790-7. doi: 10.1128/AAC.47.6.1790-1797.2003. Antimicrob Agents Chemother. 2003. PMID: 12760850 Free PMC article. - Dynamics of nasopharyngeal colonization by potential respiratory pathogens.
García-Rodríguez JA, Fresnadillo Martínez MJ. García-Rodríguez JA, et al. J Antimicrob Chemother. 2002 Dec;50 Suppl S2:59-73. doi: 10.1093/jac/dkf506. J Antimicrob Chemother. 2002. PMID: 12556435 Review. - Microbiology of bacterial respiratory infections.
Cappelletty D. Cappelletty D. Pediatr Infect Dis J. 1998 Aug;17(8 Suppl):S55-61. doi: 10.1097/00006454-199808001-00002. Pediatr Infect Dis J. 1998. PMID: 9727651 Review.
Cited by
- Alveolar macrophages in pulmonary host defence the unrecognized role of apoptosis as a mechanism of intracellular bacterial killing.
Aberdein JD, Cole J, Bewley MA, Marriott HM, Dockrell DH. Aberdein JD, et al. Clin Exp Immunol. 2013 Nov;174(2):193-202. doi: 10.1111/cei.12170. Clin Exp Immunol. 2013. PMID: 23841514 Free PMC article. Review. - Streptococcus pneumoniae and Its Virulence Factors H2O2 and Pneumolysin Are Potent Mediators of the Acute Chest Syndrome in Sickle Cell Disease.
Gonzales J, Chakraborty T, Romero M, Mraheil MA, Kutlar A, Pace B, Lucas R. Gonzales J, et al. Toxins (Basel). 2021 Feb 17;13(2):157. doi: 10.3390/toxins13020157. Toxins (Basel). 2021. PMID: 33671422 Free PMC article. Review. - Density interactions among Streptococcus pneumoniae, Haemophilus influenzae and Staphylococcus aureus in the nasopharynx of young Peruvian children.
Chien YW, Vidal JE, Grijalva CG, Bozio C, Edwards KM, Williams JV, Griffin MR, Verastegui H, Hartinger SM, Gil AI, Lanata CF, Klugman KP. Chien YW, et al. Pediatr Infect Dis J. 2013 Jan;32(1):72-7. doi: 10.1097/INF.0b013e318270d850. Pediatr Infect Dis J. 2013. PMID: 22935873 Free PMC article. - CcpA and LacD.1 affect temporal regulation of Streptococcus pyogenes virulence genes.
Kietzman CC, Caparon MG. Kietzman CC, et al. Infect Immun. 2010 Jan;78(1):241-52. doi: 10.1128/IAI.00746-09. Epub 2009 Oct 19. Infect Immun. 2010. PMID: 19841076 Free PMC article. - Characterization of colony morphology variants isolated from Streptococcus pneumoniae biofilms.
Allegrucci M, Sauer K. Allegrucci M, et al. J Bacteriol. 2007 Mar;189(5):2030-8. doi: 10.1128/JB.01369-06. Epub 2006 Dec 22. J Bacteriol. 2007. PMID: 17189375 Free PMC article.
References
- Annear D I, Dorman D C. Hydrogen peroxide accumulation during growth of the pneumococcus. Aust J Exp Bio Med Sci. 1952;30:191–195. - PubMed
- Auzat I, Chapuy-Regaud S, Le Bras G, Dos Santos D, Ogunniyi A, Le Thomas I, Garel J R, Paton J C, Trombe M C. The NADH oxidase of Streptococcus pneumoniae: its involvement in competence and virulence. Mol Microbiol. 1999;34:1018–1028. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases