The histone deacetylase-3 complex contains nuclear receptor corepressors - PubMed (original) (raw)
The histone deacetylase-3 complex contains nuclear receptor corepressors
Y D Wen et al. Proc Natl Acad Sci U S A. 2000.
Abstract
Acetylation and deacetylation of nucleosomal histones have profound effects on gene transcription in all eukaryotes. In humans, three highly homologous class I and four class II histone deacetylase (HDAC) enzymes have been identified to date. The class I deacetylases HDAC1 and HDAC2 are components of multisubunit complexes, one of which could associate with the nuclear hormone receptor corepressor, N-CoR. N-CoR also interacts with class II deacetylases HDAC4, HDAC5, and HDAC7. In comparison with HDAC1 and HDAC2, HDAC3 remains relatively uncharacterized, and very few proteins have been shown to interact with HDAC3. Using an affinity purification approach, we isolated an enzymatically active HDAC3 complex that contained members of the nuclear receptor corepressor family. Deletion analysis of N-CoR revealed that HDAC3 binds multiple N-CoR regions in vitro and that all of these regions are required for maximal binding in vivo. The N-CoR domains that interact with HDAC3 are distinct from those that bind other HDACs. Transient overexpression of HDAC3 and microinjection of Abs against HDAC3 showed that a component of transcriptional repression mediated by N-CoR depends on HDAC3. Interestingly, data suggest that interaction with a region of N-CoR augments the deacetylase activity of HDAC3. These results provide a possible molecular mechanism for HDAC3 regulation and argue that N-CoR is a platform in which distinct domains can interact with most of the known HDACs.
Figures
Figure 1
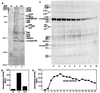
Purification of the HDAC3 complex. (A and C) Silver-stained SDS/polyacrylamide gel of the HDAC3 complex. (B and D) Histone deacetylase activity assayed from immunoaffinity-purified complexes. Each assay was performed in duplicate, and the values shown are the averages. w = wash, e = eluate, e + c = eluate and competitor.
Figure 2
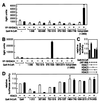
In vitro association of HDAC3 with N-CoR. (A) Schematic drawing of N-CoR. The ability of each N-CoR fusion protein to bind HDAC3 is indicated (+ or −). Representative autoradiograms of (B) in vitro translated Gal4-N-CoR protein captured by GST-HDAC3 fusion protein, (C) in vitro translated HDAC3 protein captured by GST-N-CoR fusion proteins, and (D) in vitro translated HDAC4 and HDAC5 proteins captured by GST-N-CoR fusion proteins. Several independent experiments yielded consistent results. The input lanes were loaded with one-tenth the amount of 35S-labeled proteins used in the binding reactions.
Figure 3
In vivo association of HDAC3 with N-CoR. (A and B) Mammalian two-hybrid assays were used to assess the interaction between N-CoR and HDAC3 or HDAC4 in 293 cells. VP16-HDAC3 and VP16-HDAC4 fusion proteins were expressed from pCMX-VP16-HDAC3 and pCMX-VP16-HDAC4, respectively. pUAS-p36-luc was used as the reporter. The results are the averages ± SD from at least two separate experiments. (C and D) HDAC3 enhances repression by N-CoR HDAC3-interacting domain. HDAC1, HDAC3, HDAC4, HDAC5, and various Gal4-N-CoR fusions were expressed by using plasmids described in Materials and Methods. Transfections were done in HeLa cells with the pGal4-tk-luc reporter. The results are the averages ± SD from three separate experiments.
Figure 4
(A) HDAC3 is involved in N-CoR/Pit-1-mediated repression. A reporter plasmid containing a Pit-1 response element was microinjected into 293 cells in the presence of control IgG, anti-N-CoR, or anti-HDAC3 IgG. The results were quantified as the percentage of injected cells (by rhodamine staining) that also turned blue [by 5-bromo-4-chloro-3-indolyl β-
d
-galactoside (X-Gal) staining] and shown here as the averages ± SD of two experiments performed in triplicate. (B and C) N-CoR HDAC3-interacting domain enhances deacetylase activity by HDAC3. Recombinant Flag-HDACs and Gal4-N-CoR fusion proteins were expressed in Sf9 cells, immunoprecipitated with an anti-Flag Ab. Precipitates were divided into three equal aliquots and used for deacetylase assays (B), analysis by Western blot by using the anti-Flag Ab (C Left), and analysis by Western blot by using an anti-Gal4 Ab (C Right). Deacetylase results are the averages ± SD from four separate experiments.
Similar articles
- Class II histone deacetylases: structure, function, and regulation.
Bertos NR, Wang AH, Yang XJ. Bertos NR, et al. Biochem Cell Biol. 2001;79(3):243-52. Biochem Cell Biol. 2001. PMID: 11467738 Review. - Enzymatic activity associated with class II HDACs is dependent on a multiprotein complex containing HDAC3 and SMRT/N-CoR.
Fischle W, Dequiedt F, Hendzel MJ, Guenther MG, Lazar MA, Voelter W, Verdin E. Fischle W, et al. Mol Cell. 2002 Jan;9(1):45-57. doi: 10.1016/s1097-2765(01)00429-4. Mol Cell. 2002. PMID: 11804585 - Human HDAC7 histone deacetylase activity is associated with HDAC3 in vivo.
Fischle W, Dequiedt F, Fillion M, Hendzel MJ, Voelter W, Verdin E. Fischle W, et al. J Biol Chem. 2001 Sep 21;276(38):35826-35. doi: 10.1074/jbc.M104935200. Epub 2001 Jul 20. J Biol Chem. 2001. PMID: 11466315 - The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3.
Guenther MG, Barak O, Lazar MA. Guenther MG, et al. Mol Cell Biol. 2001 Sep;21(18):6091-101. doi: 10.1128/MCB.21.18.6091-6101.2001. Mol Cell Biol. 2001. PMID: 11509652 Free PMC article. - The Role of Histone Deacetylase 3 Complex in Nuclear Hormone Receptor Action.
Ishii S. Ishii S. Int J Mol Sci. 2021 Aug 24;22(17):9138. doi: 10.3390/ijms22179138. Int J Mol Sci. 2021. PMID: 34502048 Free PMC article. Review.
Cited by
- Holocarboxylase synthetase synergizes with methyl CpG binding protein 2 and DNA methyltransferase 1 in the transcriptional repression of long-terminal repeats.
Xue J, Wijeratne SS, Zempleni J. Xue J, et al. Epigenetics. 2013 May;8(5):504-11. doi: 10.4161/epi.24449. Epub 2013 Apr 27. Epigenetics. 2013. PMID: 23624957 Free PMC article. - Histone deacetylase 1/mSin3A disrupts gamma interferon-induced CIITA function and major histocompatibility complex class II enhanceosome formation.
Zika E, Greer SF, Zhu XS, Ting JP. Zika E, et al. Mol Cell Biol. 2003 May;23(9):3091-102. doi: 10.1128/MCB.23.9.3091-3102.2003. Mol Cell Biol. 2003. PMID: 12697811 Free PMC article. - Histone deacetylase 3 (HDAC3) as an important epigenetic regulator of kidney diseases.
Zhang L, Cao W. Zhang L, et al. J Mol Med (Berl). 2022 Jan;100(1):43-51. doi: 10.1007/s00109-021-02141-8. Epub 2021 Oct 26. J Mol Med (Berl). 2022. PMID: 34698870 Review. - Regulation of SMRT corepressor dimerization and composition by MAP kinase phosphorylation.
Varlakhanova N, Hahm JB, Privalsky ML. Varlakhanova N, et al. Mol Cell Endocrinol. 2011 Jan 30;332(1-2):180-8. doi: 10.1016/j.mce.2010.10.010. Epub 2010 Oct 19. Mol Cell Endocrinol. 2011. PMID: 20965228 Free PMC article. - Posttranslational Modifications of Lipid-Activated Nuclear Receptors: Focus on Metabolism.
Becares N, Gage MC, Pineda-Torra I. Becares N, et al. Endocrinology. 2017 Feb 1;158(2):213-225. doi: 10.1210/en.2016-1577. Endocrinology. 2017. PMID: 27925773 Free PMC article. Review.
References
- Kuo M H, Allis C D. BioEssays. 1998;20:615–626. - PubMed
- Kouzarides T. Curr Opin Genet Dev. 1999;9:40–48. - PubMed
- Taunton J, Hassig C A, Schreiber S L. Science. 1996;272:408–411. - PubMed
- Yang W M, Yao Y L, Sun J M, Davie J R, Seto E. J Biol Chem. 1997;272:28001–28007. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous