Isolation and characterization of BEN, a member of the TFII-I family of DNA-binding proteins containing distinct helix-loop-helix domains - PubMed (original) (raw)
Isolation and characterization of BEN, a member of the TFII-I family of DNA-binding proteins containing distinct helix-loop-helix domains
D Bayarsaihan et al. Proc Natl Acad Sci U S A. 2000.
Abstract
The transcriptional regulation of the Hoxc8 gene is controlled during early mouse embryogenesis by an enhanceosome-like control region, termed the early enhancer (EE), located 3 kb upstream from the Hoxc8 translation start site. The EE is involved in establishing the posterior expression pattern of Hoxc8 at embryonic day (E) 8.5-9. 0. Genetic and biochemical data have shown that nuclear factors interact with this region in a sequence-specific manner. We have used a yeast one-hybrid screen in a search for transcription factors that bind to EE motifs and have isolated a novel murine DNA-binding protein, termed BEN (binding factor for early enhancer). The ORF of BEN encodes a protein of 1072 amino acids and contains six helix-loop-helix domains, a hydrophobic leucine zipper-like motif, and a serine-rich repeat. The murine BEN gene is structurally similar to the human gene TFII-I in that both genes encode unique 95-amino acid long helix-loop/span-helix domains. The BEN gene produces several major transcripts (3.6, 4.4, and 5.9 kb) present in most adult tissues and shows discrete spatial and temporal domains of expression in areas of epithelial-mesenchymal interaction during mouse embryogenesis from E9.5 to E12.5. Several BEN-encoded polypeptides of different sizes ranging from 165 to 40 kDa were identified by Western blot analysis using BEN-specific polyclonal Abs. We propose, on the bases of sequence homology, that BEN is the mouse ortholog of the recently described human gene, WBSCR11, known also as GTF2IRD1, GTF3, Cream1, and MusTRD1. This gene is deleted hemizygously in individuals with Williams Syndrome, an autosomal dominant genetic condition characterized by complex physical, cognitive, and behavioral traits resulting from a perturbed developmental process.
Figures
Figure 1
Nucleotide, deduced amino acid sequence, and structural organization of BEN. (A) The nucleotide and amino acid sequence of BEN. The six HLH repeat domains are highlighted in black. The putative nuclear localization signal is double underlined and the serine-rich region is single underlined. The amino acid residues in the hydrophobic leucine-zipper motif are indicated by circles. The mRNA destabilization motif is marked by asterisks. Two polyadenylation-like sequences are marked by dots. (B) Domain organization of BEN. LZ, potential leucine zipper motif; R1–R6, HLH repeat domains; NLS, putative nuclear localization signal; and SR, serine-rich region.
Figure 2
Sequence alignments of BEN and TFII-I. (A) Amino acid sequence alignments of the HLH repeat domains of BEN and TFII-I (R1–R6). Conserved residues are highlighted with black, and consensus amino acid sequences are indicated below. (B) Alignment of the N-terminal 59-aa residues of BEN (amino acids 30–88) and TFII-I (amino acids 22–80).
Figure 3
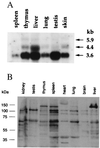
BEN expression in mouse tissues. (A) BEN mRNA was detected by Northern blot analysis of mRNA from adult mouse tissues (2 μg per lane) with BEN cDNA probe. The positions of RNA molecular weight markers are shown on the left. (B) Immunodetection of the BEN protein in mouse tissues. Western blot analysis was performed with BEN-specific polyclonal Abs. Total cellular extracts were run on 7.5% SDS/PAGE gel and processed as described in Materials and Methods. The protein standards (Novagene) are shown on the left.
Figure 4
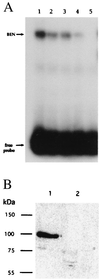
DNA-binding studies of BEN. (A) EMSA of the binding of in vitro translated BEN protein to the EFG probe. All lanes contain labeled EFG oligonucleotide and in vitro translated BEN protein. Lane 1, no competitor; lane 2 to 5 have 5-, 10-, 20-, and 40-fold excess of cold EFG oligonucleotide. (B) Southwestern blot analysis of BEN shows that affinity-purified His-tagged BEN binds the EFG sequence. The proteins purified on a nickel column were run on 10% SDS/PAGE and electroblotted to a nitrocellulose membrane. The filters were probed with labeled EFG (lane 1) or UCD (lane 2) oligonucleotides, respectively. The single band around 90 kDa is the truncated His-tagged BEN fusion protein.
Similar articles
- A transcription factor involved in skeletal muscle gene expression is deleted in patients with Williams syndrome.
Tassabehji M, Carette M, Wilmot C, Donnai D, Read AP, Metcalfe K. Tassabehji M, et al. Eur J Hum Genet. 1999 Oct-Nov;7(7):737-47. doi: 10.1038/sj.ejhg.5200396. Eur J Hum Genet. 1999. PMID: 10573005 - Isolation and characterisation of GTF2IRD2, a novel fusion gene and member of the TFII-I family of transcription factors, deleted in Williams-Beuren syndrome.
Tipney HJ, Hinsley TA, Brass A, Metcalfe K, Donnai D, Tassabehji M. Tipney HJ, et al. Eur J Hum Genet. 2004 Jul;12(7):551-60. doi: 10.1038/sj.ejhg.5201174. Eur J Hum Genet. 2004. PMID: 15100712 - GTF2IRD2 is located in the Williams-Beuren syndrome critical region 7q11.23 and encodes a protein with two TFII-I-like helix-loop-helix repeats.
Makeyev AV, Erdenechimeg L, Mungunsukh O, Roth JJ, Enkhmandakh B, Ruddle FH, Bayarsaihan D. Makeyev AV, et al. Proc Natl Acad Sci U S A. 2004 Jul 27;101(30):11052-7. doi: 10.1073/pnas.0404150101. Epub 2004 Jul 8. Proc Natl Acad Sci U S A. 2004. PMID: 15243160 Free PMC article. - Identification of a novel slow-muscle-fiber enhancer binding protein, MusTRD1.
O'Mahoney JV, Guven KL, Lin J, Joya JE, Robinson CS, Wade RP, Hardeman EC. O'Mahoney JV, et al. Mol Cell Biol. 1998 Nov;18(11):6641-52. doi: 10.1128/MCB.18.11.6641. Mol Cell Biol. 1998. PMID: 9774679 Free PMC article.
Cited by
- Regulation of RNA Polymerase II Transcription Initiation and Elongation by Transcription Factor TFII-I.
Linzer N, Trumbull A, Nar R, Gibbons MD, Yu DT, Strouboulis J, Bungert J. Linzer N, et al. Front Mol Biosci. 2021 May 13;8:681550. doi: 10.3389/fmolb.2021.681550. eCollection 2021. Front Mol Biosci. 2021. PMID: 34055891 Free PMC article. Review. - Functions of Gtf2i and Gtf2ird1 in the developing brain: transcription, DNA binding and long-term behavioral consequences.
Kopp ND, Nygaard KR, Liu Y, McCullough KB, Maloney SE, Gabel HW, Dougherty JD. Kopp ND, et al. Hum Mol Genet. 2020 Jun 3;29(9):1498-1519. doi: 10.1093/hmg/ddaa070. Hum Mol Genet. 2020. PMID: 32313931 Free PMC article. - The developmental and genetic basis of 'clubfoot' in the peroneal muscular atrophy mutant mouse.
Collinson JM, Lindström NO, Neves C, Wallace K, Meharg C, Charles RH, Ross ZK, Fraser AM, Mbogo I, Oras K, Nakamoto M, Barker S, Duce S, Miedzybrodzka Z, Vargesson N. Collinson JM, et al. Development. 2018 Feb 8;145(3):dev160093. doi: 10.1242/dev.160093. Development. 2018. PMID: 29439133 Free PMC article. - Structural variants in genes associated with human Williams-Beuren syndrome underlie stereotypical hypersociability in domestic dogs.
vonHoldt BM, Shuldiner E, Koch IJ, Kartzinel RY, Hogan A, Brubaker L, Wanser S, Stahler D, Wynne CDL, Ostrander EA, Sinsheimer JS, Udell MAR. vonHoldt BM, et al. Sci Adv. 2017 Jul 19;3(7):e1700398. doi: 10.1126/sciadv.1700398. eCollection 2017 Jul. Sci Adv. 2017. PMID: 28776031 Free PMC article. - The nuclear localization pattern and interaction partners of GTF2IRD1 demonstrate a role in chromatin regulation.
Carmona-Mora P, Widagdo J, Tomasetig F, Canales CP, Cha Y, Lee W, Alshawaf A, Dottori M, Whan RM, Hardeman EC, Palmer SJ. Carmona-Mora P, et al. Hum Genet. 2015 Oct;134(10):1099-115. doi: 10.1007/s00439-015-1591-0. Epub 2015 Aug 15. Hum Genet. 2015. PMID: 26275350
References
- Krumlauf R. Cell. 1994;78:191–201. - PubMed
- McGinnis W R, Garber J W, Kuroiwa A, Gehring W. Cell. 1984;37:403–408. - PubMed
- Ruddle F H, Bartels J L, Bentley K L, Kappen C, Murtha M T, Pendleton J W. Annu Rev Genet. 1994;28:432–442. - PubMed
- Shashikant C S, Bieberich C J, Belting H G, Wang C H, Borbely M A, Ruddle F H. Development. 1995;121:4339–4347. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases