Selective membrane permeabilization by the rotavirus VP5* protein is abrogated by mutations in an internal hydrophobic domain - PubMed (original) (raw)
Selective membrane permeabilization by the rotavirus VP5* protein is abrogated by mutations in an internal hydrophobic domain
W Dowling et al. J Virol. 2000 Jul.
Abstract
Rotavirus infectivity is dependent on the proteolytic cleavage of the VP4 spike protein into VP8* and VP5* proteins. Proteolytically activated virus, as well as expressed VP5*, permeabilizes membranes, suggesting that cleavage exposes a membrane-interactive domain of VP5* which effects rapid viral entry. The VP5* protein contains a single long hydrophobic domain (VP5*-HD, residues 385 to 404) at an internal site. In order to address the role of the VP5*-HD in permeabilizing cellular membranes, we analyzed the entry of o-nitrophenyl-beta-D-galactopyranoside (ONPG) into cells induced to express VP5* or mutated VP5* polypeptides. Following IPTG (isopropyl-beta-D-thiogalactopyranoside) induction, VP5* and VP5* truncations containing the VP5*-HD permeabilized cells to the entry and cleavage of ONPG, while VP8* and control proteins had no effect on cellular permeability. Expression of VP5* deletions containing residues 265 to 474 or 265 to 404 permeabilized cells; however, C-terminal truncations which remove the conserved GGA (residues 399 to 401) within the HD abolished membrane permeability. Site-directed mutagenesis of the VP5-HD further demonstrated a requirement for residues within the HD for VP5*-induced membrane permeability. Functional analysis of mutant VP5*s indicate that conserved glycines within the HD are required and suggest that a random coiled structure rather than the strictly hydrophobic character of the domain is required for permeability. Expressed VP5* did not alter bacterial growth kinetics or lyse bacteria following induction. Instead, VP5*-mediated size-selective membrane permeability, releasing 376-Da carboxyfluorescein but not 4-kDa fluorescein isothiocyanate-dextran from preloaded liposomes. These findings suggest that the fundamental role for VP5* in the rotavirus entry process may be to expose triple-layered particles to low [Ca](i), which uncoats the virus, rather than to effect the detergent-like lysis of early endosomal membranes.
Figures
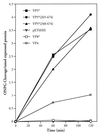
FIG. 1
VP5* induction permeabilizes bacteria. Bacterial permeabilization following IPTG induction was measured as previously described (12). E. coli BL21(DE3) cells containing pET-6HIS plasmids expressing VP4, VP8*, VP5*, VP5*(248–474), VP5*(265–474), or empty controls were grown to an OD600 of 0.2 to 0.4 and induced by the addition of IPTG (1 mM). At the indicated times cells were pelleted, resuspended in LB containing 50 μg of streptomycin per ml and 2 mM ONPG and incubated at 30°C for 10 min (26). Cleaved ONPG in supernatants was assayed spectrophotometrically (OD420), and the absorbance was standardized to nanomoles of recombinant expressed protein. This experiment was repeated six times with similar results.
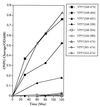
FIG. 2
Permeabilizing activity of VP5* truncations. E. coli BL21(DE3) containing pET-6HIS plasmids expressing the indicated VP5* truncations were grown and induced as previously described (12). Bacterial permeabilization was determined by using the ONPG cleavage assay as in Fig. 1 and standardized to OD600 of the culture at each time point. Uninduced controls resulted in ONPG cleavage of <0.1/OD600. This experiment was repeated at least three times with similar results.
FIG. 3
Mutagenesis of the VP5* HD inhibits membrane permeability. (A) VP5*(265–474) or indicated VP5*(265–474) point mutants were assayed for their ability to permeabilize E. coli as measured by the ONPG cleavage assay (Fig. 1). Bacteria were grown, IPTG induced, and assayed for ONPG cleavage as in Fig. 1. Cleaved ONPG in the supernatants was assayed as in Fig. 2. Each mutant was tested at least three times with similar results. (B) Summary of VP5* mutants. The permeability induced by oligo-nucleotide-directed VP5* mutants is summarized. Residues and introduced changes within the VP5* hydrophobic domain mutants are shown along with their ability (+) or inability (−) to permeabilize cells. Residues highly conserved among rotavirus strains are indicated in boldface. ∗, residue change in neutralizing MAb escape mutants.
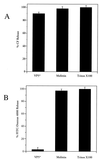
FIG. 4
Bacteria are not lysed by VP5* expression. (A) β-Galactosidase is not released following VP5* expression. E. coli BL21(DE3)/pLysS containing the pET-6HIS plasmid with no insert or expressing either VP5*(265–474) or VP5*(248–776) were assayed for their ability to cause the release of β-galactosidase into the culture medium as measured by ONPG cleavage. Bacteria were grown to an OD600 of 0.2 to 0.4 and induced by the addition of IPTG (1 mM). Samples of each culture were removed at the indicated times after IPTG induction, and the OD600 was measured. Cells were pelleted, and both resuspended cells and supernatants were analyzed for ONPG cleavage as in Fig. 1. This experiment was repeated two times with similar results. (B) VP5* expression does not alter bacterial growth. E. coli BL21(DE3)/pLysS transformed with pET-6HIS plasmids expressing either VP5*(265–474), VP5*, or VP8* were grown to an OD600 of 0.2 to 0.4 and split into separate tubes. One tube was induced by the addition of IPTG (1 mM), and another was grown in parallel as a control. At given times postinduction, samples of each culture were removed and the OD600 was measured; growth curves of induced and uninduced cultures are shown. This experiment was repeated three times with similar results.
FIG. 5
VP5* selectively permeabilizes liposomes. Purified LUVs containing either CF (70 mM) or FITC-dextran 4000 (FD-4) (30 mM) were incubated in 1.6 ml of TN buffer at 37°C for 30 min with nickel affinity-purified VP5* (4 μg, 44 nM) or melittin (0.03 nM) in duplicate (31, 42). (A) To measure CF release, fluorescence dequenching of samples was measured in a fluorimeter at an excitation wavelength of 490 nm and an emission wavelength of 520 nm. The total CF content of each sample was assessed by the addition of Triton X-100 (0.125%) and quantitated as described in Materials and Methods. (B) FD-4 is not present at self-quenching concentrations, and release was determined after centrifuging samples in a Centricon-100 microseparator (Amicon) and measuring released FD-4 in the effluent using a Perkin-Elmer Luminescence Spectrometer (LS-5B) at 520 nm (490-nm excitation). Liposome-associated FD-4 was assayed after resuspension in 1.6 ml of TN buffer plus 0.125% Triton X-100. Released FD-4 is expressed as a percentage of the total fluorescence (the sum of the effluent and the retentate). This experiment was repeated twice with similar results.
FIG. 6
Role of VP5* in rotavirus entry. The role of VP5* in the rotavirus entry process is hypothesized in light of its ability to selectively permeabilize membranes. Rotavirus is drawn as a circle with protruding VP4 spikes initially entering an early endosome. The outer capsid proteins on the virion are presumed to maintain contacts with receptors, while fusogenic domains of VP5* are exposed following VP4 cleavage in order to selectively permeabilize the early endosomal membrane. Permeabilization of early endosomes presumably lowers the [Ca]i surrounding the virions and permits virion uncoating.
Similar articles
- Discrete domains within the rotavirus VP5* direct peripheral membrane association and membrane permeability.
Golantsova NE, Gorbunova EE, Mackow ER. Golantsova NE, et al. J Virol. 2004 Feb;78(4):2037-44. doi: 10.1128/jvi.78.4.2037-2044.2004. J Virol. 2004. PMID: 14747568 Free PMC article. - Rotavirus capsid protein VP5* permeabilizes membranes.
Denisova E, Dowling W, LaMonica R, Shaw R, Scarlata S, Ruggeri F, Mackow ER. Denisova E, et al. J Virol. 1999 Apr;73(4):3147-53. doi: 10.1128/JVI.73.4.3147-3153.1999. J Virol. 1999. PMID: 10074166 Free PMC article. - The VP5 domain of VP4 can mediate attachment of rotaviruses to cells.
Zárate S, Espinosa R, Romero P, Méndez E, Arias CF, López S. Zárate S, et al. J Virol. 2000 Jan;74(2):593-9. doi: 10.1128/jvi.74.2.593-599.2000. J Virol. 2000. PMID: 10623720 Free PMC article. - Rotavirus protein expression is important for virus assembly and pathogenesis.
Tian P, Ball JM, Zeng CQ, Estes MK. Tian P, et al. Arch Virol Suppl. 1996;12:69-77. doi: 10.1007/978-3-7091-6553-9_8. Arch Virol Suppl. 1996. PMID: 9015103 Review.
Cited by
- Rotavirus VP8*: phylogeny, host range, and interaction with histo-blood group antigens.
Liu Y, Huang P, Tan M, Liu Y, Biesiada J, Meller J, Castello AA, Jiang B, Jiang X. Liu Y, et al. J Virol. 2012 Sep;86(18):9899-910. doi: 10.1128/JVI.00979-12. Epub 2012 Jul 3. J Virol. 2012. PMID: 22761376 Free PMC article. - Entry of Bluetongue Virus Capsid Requires the Late Endosome-specific Lipid Lysobisphosphatidic Acid.
Patel A, Mohl BP, Roy P. Patel A, et al. J Biol Chem. 2016 Jun 3;291(23):12408-19. doi: 10.1074/jbc.M115.700856. Epub 2016 Apr 1. J Biol Chem. 2016. PMID: 27036941 Free PMC article. - Genomic Analysis of G2P[4] Group A Rotaviruses in Zambia Reveals Positive Selection in Amino Acid Site 7 of Viral Protein 3.
Mwangi PN, Potgieter RL, Simwaka J, Mpabalwani EM, Mwenda JM, Mogotsi MT, Magagula N, Esona MD, Steele AD, Seheri ML, Nyaga MM. Mwangi PN, et al. Viruses. 2023 Feb 11;15(2):501. doi: 10.3390/v15020501. Viruses. 2023. PMID: 36851715 Free PMC article. - Rotavirus gene silencing by small interfering RNAs.
Déctor MA, Romero P, López S, Arias CF. Déctor MA, et al. EMBO Rep. 2002 Dec;3(12):1175-80. doi: 10.1093/embo-reports/kvf234. Epub 2002 Nov 21. EMBO Rep. 2002. PMID: 12446562 Free PMC article. - Generation of recombinant rotavirus with an antigenic mosaic of cross-reactive neutralization epitopes on VP4.
Komoto S, Kugita M, Sasaki J, Taniguchi K. Komoto S, et al. J Virol. 2008 Jul;82(13):6753-7. doi: 10.1128/JVI.00601-08. Epub 2008 Apr 23. J Virol. 2008. PMID: 18434390 Free PMC article.
References
- Beaumelle B, Bensammar L, Bienvenue A. Selective translocation of the A chain diphtheria toxin across the membrane of purified endosomes. J Biol Chem. 1992;267:11525–11531. - PubMed
- Butko P, Huang F, Pusztai-Carey M, Surewicz W K. Membrane permeabilization induced by cytolytic delta-endotoxin CytA from Bacillus thuringiensis var. israelensis. Biochemistry. 1996;35:11355–11360. - PubMed
- Charpilienne A, Abad M J, Michelangeli F, Alvarado F, Vasseur M, Cohen J, Ruiz M C. Solubilized and cleaved VP7, the outer glycoprotein of rotavirus, induces permeabilization of cell membrane vesicles. J Gen Virol. 1997;78:1367–1371. - PubMed
- Ciarlet M, Estes M K. Human and most animal rotavirus strains do not require the presence of sialic acid on the cell surface for efficient infectivity. J Gen Virol. 1999;80:943–948. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources