Mechanism of transcriptional regulation by methyl-CpG binding protein MBD1 - PubMed (original) (raw)
Mechanism of transcriptional regulation by methyl-CpG binding protein MBD1
N Fujita et al. Mol Cell Biol. 2000 Jul.
Abstract
MBD1 is a mammalian protein that binds symmetrically methylated CpG sequences and regulates gene expression in association with DNA methylation. This protein possesses a conserved sequence, named methyl-CpG binding domain (MBD), among a family of methyl-CpG binding proteins that mediate the biological consequences of the methylation. In addition, MBD1 has at least five isoforms due to alternative splicing events, resulting in the presence of CXXC1, CXXC2, and CXXC3 in MBD1 isoforms v1 (MBD1v1) and MBD1v2, and CXXC1 and CXXC2 in MBD1v3 and -v4. In the present study, we have investigated the significance of MBD, CXXC, and the C-terminal transcriptional repression domain (TRD) in MBD1. A bacterially expressed MBD binds efficiently to densely methylated rather than to sparsely methylated DNAs. In both methylation-deficient Drosophila melanogaster SL2 cells and mammalian CHO-K1 cells, MBD1v1 represses transcription preferentially from both unmethylated and sparsely methylated promoters, while MBD1v3 inhibits densely methylated but not unmethylated promoter activities. The CXXC3 sequence in MBD1v1 is responsible for the ability to bind unmethylated promoter. Furthermore, we have constructed mutant-type MBD1s in which the functionally important residues Arg22, Arg30, Asp32, Tyr34, Arg44, Ser45, and Tyr52 are changed to alanine to investigate the correlation between the structure and function of the MBD in MBD1. Excepting those for Ser45 and Tyr52, none of the recombinant MBD mutants bound to the densely methylated or unmethylated DNAs, and green fluorescent protein-fused MBD1 mutants did not localize properly in the nucleus. All the MBD1v1 and -v3 mutants lost the activity of methylation-dependent gene repression. Based on these findings we have concluded that MBD1 acts as a transcriptional regulator depending on the density of methyl-CpG pairs through the cooperation of MBD, CXXC, and TRD sequences.
Figures
FIG. 1
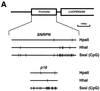
Effect of MBD1 isoforms on methylated and unmethylated promoters. (A) PCR-amplified DNA fragments from human imprinted SNRPN and tumor suppressor p16 genes were used for a band shift analysis and subcloned upstream of a luciferase cDNA in a pGL3-Basic vector. The PCR fragments and pGL3 constructs were methylated in vitro using _Hpa_II, _Hha_I, and _Sss_I (CpG) methyltransferases. The methyl-CpG sites modified by these enzymes are shown by vertical lines. (B) Band shift of methylated DNA complexed with the methyl-CpG binding domain of MBD1. Unmethylated (−) and methylated fragments containing SNRPN promoter were incubated with MBD1 (residues 1 to 75) or GST. In the upper and lower panels, the amount of the protein incubated with DNA fragments was 0.5 and 1.0 μg, respectively. (C and D) Regulation of Sp1-activated transcription by MBD1v1 and -v3 in Drosophila SL2 cells (SNRPN [C] or p16 [D]). Unmethylated (M-) or _Hpa_II-, _Hha_I-, or _Sss_I-methylated promoter-inserted pGL3 vector (0.5 μg) was cotransfected with Sp1-expressing plasmid pPacSp1 (0.5 μg), MBD1-expressing plasmids (pAc5.1-MBD1v1 and pAc5.1-MBD1v3) (0 to 1.0 μg), and insertless plasmid pAc5.1/V5-His (mock) (1.0 to 0 μg). The luciferase activity of unmethylated pGL3 in combination with pPacSp1 and 1.0 μg of pAc5.1/V5-His (mock) was normalized to 100, and the relative luciferase activities (means + standard deviations [error bars]) were determined after correcting the transfection efficiency by pAc5.1-pRL (0.1 μg). (E) Detection of endogenous MBD1 by an antibody raised against the recombinant MBD1. MBD1 was found to be approximately 80 kDa in HeLa and A549 cells but not in SL2 and CHO-K1 cells.
FIG. 1
Effect of MBD1 isoforms on methylated and unmethylated promoters. (A) PCR-amplified DNA fragments from human imprinted SNRPN and tumor suppressor p16 genes were used for a band shift analysis and subcloned upstream of a luciferase cDNA in a pGL3-Basic vector. The PCR fragments and pGL3 constructs were methylated in vitro using _Hpa_II, _Hha_I, and _Sss_I (CpG) methyltransferases. The methyl-CpG sites modified by these enzymes are shown by vertical lines. (B) Band shift of methylated DNA complexed with the methyl-CpG binding domain of MBD1. Unmethylated (−) and methylated fragments containing SNRPN promoter were incubated with MBD1 (residues 1 to 75) or GST. In the upper and lower panels, the amount of the protein incubated with DNA fragments was 0.5 and 1.0 μg, respectively. (C and D) Regulation of Sp1-activated transcription by MBD1v1 and -v3 in Drosophila SL2 cells (SNRPN [C] or p16 [D]). Unmethylated (M-) or _Hpa_II-, _Hha_I-, or _Sss_I-methylated promoter-inserted pGL3 vector (0.5 μg) was cotransfected with Sp1-expressing plasmid pPacSp1 (0.5 μg), MBD1-expressing plasmids (pAc5.1-MBD1v1 and pAc5.1-MBD1v3) (0 to 1.0 μg), and insertless plasmid pAc5.1/V5-His (mock) (1.0 to 0 μg). The luciferase activity of unmethylated pGL3 in combination with pPacSp1 and 1.0 μg of pAc5.1/V5-His (mock) was normalized to 100, and the relative luciferase activities (means + standard deviations [error bars]) were determined after correcting the transfection efficiency by pAc5.1-pRL (0.1 μg). (E) Detection of endogenous MBD1 by an antibody raised against the recombinant MBD1. MBD1 was found to be approximately 80 kDa in HeLa and A549 cells but not in SL2 and CHO-K1 cells.
FIG. 1
Effect of MBD1 isoforms on methylated and unmethylated promoters. (A) PCR-amplified DNA fragments from human imprinted SNRPN and tumor suppressor p16 genes were used for a band shift analysis and subcloned upstream of a luciferase cDNA in a pGL3-Basic vector. The PCR fragments and pGL3 constructs were methylated in vitro using _Hpa_II, _Hha_I, and _Sss_I (CpG) methyltransferases. The methyl-CpG sites modified by these enzymes are shown by vertical lines. (B) Band shift of methylated DNA complexed with the methyl-CpG binding domain of MBD1. Unmethylated (−) and methylated fragments containing SNRPN promoter were incubated with MBD1 (residues 1 to 75) or GST. In the upper and lower panels, the amount of the protein incubated with DNA fragments was 0.5 and 1.0 μg, respectively. (C and D) Regulation of Sp1-activated transcription by MBD1v1 and -v3 in Drosophila SL2 cells (SNRPN [C] or p16 [D]). Unmethylated (M-) or _Hpa_II-, _Hha_I-, or _Sss_I-methylated promoter-inserted pGL3 vector (0.5 μg) was cotransfected with Sp1-expressing plasmid pPacSp1 (0.5 μg), MBD1-expressing plasmids (pAc5.1-MBD1v1 and pAc5.1-MBD1v3) (0 to 1.0 μg), and insertless plasmid pAc5.1/V5-His (mock) (1.0 to 0 μg). The luciferase activity of unmethylated pGL3 in combination with pPacSp1 and 1.0 μg of pAc5.1/V5-His (mock) was normalized to 100, and the relative luciferase activities (means + standard deviations [error bars]) were determined after correcting the transfection efficiency by pAc5.1-pRL (0.1 μg). (E) Detection of endogenous MBD1 by an antibody raised against the recombinant MBD1. MBD1 was found to be approximately 80 kDa in HeLa and A549 cells but not in SL2 and CHO-K1 cells.
FIG. 1
Effect of MBD1 isoforms on methylated and unmethylated promoters. (A) PCR-amplified DNA fragments from human imprinted SNRPN and tumor suppressor p16 genes were used for a band shift analysis and subcloned upstream of a luciferase cDNA in a pGL3-Basic vector. The PCR fragments and pGL3 constructs were methylated in vitro using _Hpa_II, _Hha_I, and _Sss_I (CpG) methyltransferases. The methyl-CpG sites modified by these enzymes are shown by vertical lines. (B) Band shift of methylated DNA complexed with the methyl-CpG binding domain of MBD1. Unmethylated (−) and methylated fragments containing SNRPN promoter were incubated with MBD1 (residues 1 to 75) or GST. In the upper and lower panels, the amount of the protein incubated with DNA fragments was 0.5 and 1.0 μg, respectively. (C and D) Regulation of Sp1-activated transcription by MBD1v1 and -v3 in Drosophila SL2 cells (SNRPN [C] or p16 [D]). Unmethylated (M-) or _Hpa_II-, _Hha_I-, or _Sss_I-methylated promoter-inserted pGL3 vector (0.5 μg) was cotransfected with Sp1-expressing plasmid pPacSp1 (0.5 μg), MBD1-expressing plasmids (pAc5.1-MBD1v1 and pAc5.1-MBD1v3) (0 to 1.0 μg), and insertless plasmid pAc5.1/V5-His (mock) (1.0 to 0 μg). The luciferase activity of unmethylated pGL3 in combination with pPacSp1 and 1.0 μg of pAc5.1/V5-His (mock) was normalized to 100, and the relative luciferase activities (means + standard deviations [error bars]) were determined after correcting the transfection efficiency by pAc5.1-pRL (0.1 μg). (E) Detection of endogenous MBD1 by an antibody raised against the recombinant MBD1. MBD1 was found to be approximately 80 kDa in HeLa and A549 cells but not in SL2 and CHO-K1 cells.
FIG. 1
Effect of MBD1 isoforms on methylated and unmethylated promoters. (A) PCR-amplified DNA fragments from human imprinted SNRPN and tumor suppressor p16 genes were used for a band shift analysis and subcloned upstream of a luciferase cDNA in a pGL3-Basic vector. The PCR fragments and pGL3 constructs were methylated in vitro using _Hpa_II, _Hha_I, and _Sss_I (CpG) methyltransferases. The methyl-CpG sites modified by these enzymes are shown by vertical lines. (B) Band shift of methylated DNA complexed with the methyl-CpG binding domain of MBD1. Unmethylated (−) and methylated fragments containing SNRPN promoter were incubated with MBD1 (residues 1 to 75) or GST. In the upper and lower panels, the amount of the protein incubated with DNA fragments was 0.5 and 1.0 μg, respectively. (C and D) Regulation of Sp1-activated transcription by MBD1v1 and -v3 in Drosophila SL2 cells (SNRPN [C] or p16 [D]). Unmethylated (M-) or _Hpa_II-, _Hha_I-, or _Sss_I-methylated promoter-inserted pGL3 vector (0.5 μg) was cotransfected with Sp1-expressing plasmid pPacSp1 (0.5 μg), MBD1-expressing plasmids (pAc5.1-MBD1v1 and pAc5.1-MBD1v3) (0 to 1.0 μg), and insertless plasmid pAc5.1/V5-His (mock) (1.0 to 0 μg). The luciferase activity of unmethylated pGL3 in combination with pPacSp1 and 1.0 μg of pAc5.1/V5-His (mock) was normalized to 100, and the relative luciferase activities (means + standard deviations [error bars]) were determined after correcting the transfection efficiency by pAc5.1-pRL (0.1 μg). (E) Detection of endogenous MBD1 by an antibody raised against the recombinant MBD1. MBD1 was found to be approximately 80 kDa in HeLa and A549 cells but not in SL2 and CHO-K1 cells.
FIG. 2
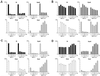
Transcriptional regulation by MBD1 isoforms and their mutants with deletions of the methyl-CpG binding domain in mammalian CHO-K1 cells. Unmethylated or _Hpa_II-, _Hha_I-, or _Sss_I-methylated promoter-inserted pGL3 vector (0.5 μg) was cotransfected with MBD1-expressing plasmids (pCGN-MBD1v1, pCGN-MBD1v3, pCGN-MBD1v1ΔN, and pCGN-MBD1v3ΔN) (0 to 1.0 μg) and insertless plasmid pCGN (mock) (1.0 to 0 μg). The luciferase activity of unmethylated pGL3 in combination with 1.0 μg of pCGN (mock) was normalized to 10,000, and the relative luciferase activities (means + standard deviations [error bars]) were determined after correcting the transfection efficiency by pRL-SV40 (0.1 μg). The combinations of pGL3-SNRPN and pCGN-MBD1v1 or pCGN-MBD1v1ΔN (A), pGL3-SNRPN and pCGN-MBD1v3 or pCGN-MBD1v3ΔN (B), pGL3-p16 and pCGN-MBD1v1 or pCGN-MBD1v1ΔN (C), and pGL3-p16 and pCGN-MBD1v3 or pCGN-MBD1v3ΔN (D) are shown.
FIG. 3
Effect of MBD1v1 mutants deleted of the CXXC domains on Sp1-activated transcription in Drosophila SL2 cells. (A) Four pAc5.1/V5-His plasmids expressing deletion mutants of MBD1v1 were designated MBD1v1Δ1 to MBD1v1Δ4. (B) Each of the plasmids for MBD1v1 deletion mutants (1.0 μg) was transfected into Drosophila cells together with pPacSp1 (0.5 μg), and either unmethylated (black bars) or _Hpa_II (gray bars)-, _Hha_I (white bars)-, or _Sss_I (hatched bars)-methylated pGL3-SNRPN (0.5 μg). The luciferase activity of unmethylated pGL3-SNRPN in combination with pPacSp1 and 1.0 μg of pAc5.1/V5-His (mock) was normalized to 100. Relative luciferase activities (means + standard deviations [error bars]) are given.
FIG. 3
Effect of MBD1v1 mutants deleted of the CXXC domains on Sp1-activated transcription in Drosophila SL2 cells. (A) Four pAc5.1/V5-His plasmids expressing deletion mutants of MBD1v1 were designated MBD1v1Δ1 to MBD1v1Δ4. (B) Each of the plasmids for MBD1v1 deletion mutants (1.0 μg) was transfected into Drosophila cells together with pPacSp1 (0.5 μg), and either unmethylated (black bars) or _Hpa_II (gray bars)-, _Hha_I (white bars)-, or _Sss_I (hatched bars)-methylated pGL3-SNRPN (0.5 μg). The luciferase activity of unmethylated pGL3-SNRPN in combination with pPacSp1 and 1.0 μg of pAc5.1/V5-His (mock) was normalized to 100. Relative luciferase activities (means + standard deviations [error bars]) are given.
FIG. 4
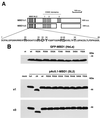
Site-directed mutagenesis of the methyl-CpG binding domain in MBD1. (A) Diagram of MBD1v1 and -v3. MBD1 has an MBD, an NLS, and two or three cysteine-rich CXXC domains due to alternative splicing events. Seven amino acid residues indicated by oversize capital letters are important for the methylated DNA binding, and they were mutated to alanine. The numbers above the residues indicate the positions relative to the N terminus. a.a., amino acids. (B) Expression of wild-type (wt) and mutant MBD1v1 and MBD1v3 in HeLa and Drosophila SL2 cells. pEGFP-wt. MBD1 and pEGFP-MBD1(R22A) to pEGFP-MBD1(Y52A) express wild-type and mutant MBD1 fused to GFP in HeLa cells (upper panel). In SL2 cells, pAc5.1-MBD1v1 and pAc5.1-MBD1v3 express full-length MBD1v1 and MBD1v3, while pAc5.1-MBD1v1ΔN and pAc5.1-MBD1v3ΔN express MBD1v1 and MBD1v3 with the MBD (residues 1 to 61) deleted, respectively. pAc5.1-wt. MBD1v1, pAc5.1-wt. MBD1v3, pAc5.1-MBD1v1(R22A) to pAc5.1-MBD1v1(Y52A), and pAc5.1-MBD1v3(R22A) to pAc5.1-MBD1v3(Y52A) express wild-type and mutant MBD1 (lower panel). A Western blot analysis was performed using anti-GFP and anti-MBD1 polyclonal antibodies. The lysates from nontransfected cells (nt) and mock-transfected cells (mock) are used as a control.
FIG. 5
Residues within the methyl-CpG binding domain of MBD1 required for the methylated DNA binding, intranuclear localization, and transcriptional repression of methylated promoter. (A) Band shift of methylated DNA complexed with wild-type (wt) and mutant MBD1. Unmethylated (−) and _Sss_I-methylated (+) DNA fragments of the SNRPN promoter were incubated with MBD1 (residues 1 to 75) or GST. (B) Intranuclear localization of GFP-fused MBD1 (full-length and N-terminal deletion ΔN) and MBD1 (residues 1 to 89) with the above-mentioned point mutations. (C) _Hha_I- or _Sss_I-methylated pGL3-SNRPN (0.5 μg) was cotransfected with pPacSp1 (0.5 μg) and one of the MBD1-expressing plasmids [pAc5.1-MBD1v1, pAc5.1-MBD1v1ΔN, pAc5.1-wt. MBD1v1, pAc5.1-MBD1v1(R22A) to pAc5.1-MBD1v1(Y52A), pAc5.1-MBD1v3, pAc5.1-MBD1v3ΔN, pAc5.1-wt. MBD1v3, and pAc5.1-MBD1v3(R22A) to pAc5.1-MBD1v3(Y52A)] (1.0 μg) or insertless plasmid pAc5.1/V5-His (mock) (1.0 μg). The luciferase activity of unmethylated pGL3-SNRPN in combination with pPacSp1 and pAc5.1/V5-His (mock) was normalized to 100 (data not shown). Relative luciferase activities (means + standard deviations) are given. The numbers 22, 30, 32, 34, 44, 45, and 52 correspond to R22A, R30A, D32A, Y34A, R44A, S45A, and Y52A, respectively.
FIG. 5
Residues within the methyl-CpG binding domain of MBD1 required for the methylated DNA binding, intranuclear localization, and transcriptional repression of methylated promoter. (A) Band shift of methylated DNA complexed with wild-type (wt) and mutant MBD1. Unmethylated (−) and _Sss_I-methylated (+) DNA fragments of the SNRPN promoter were incubated with MBD1 (residues 1 to 75) or GST. (B) Intranuclear localization of GFP-fused MBD1 (full-length and N-terminal deletion ΔN) and MBD1 (residues 1 to 89) with the above-mentioned point mutations. (C) _Hha_I- or _Sss_I-methylated pGL3-SNRPN (0.5 μg) was cotransfected with pPacSp1 (0.5 μg) and one of the MBD1-expressing plasmids [pAc5.1-MBD1v1, pAc5.1-MBD1v1ΔN, pAc5.1-wt. MBD1v1, pAc5.1-MBD1v1(R22A) to pAc5.1-MBD1v1(Y52A), pAc5.1-MBD1v3, pAc5.1-MBD1v3ΔN, pAc5.1-wt. MBD1v3, and pAc5.1-MBD1v3(R22A) to pAc5.1-MBD1v3(Y52A)] (1.0 μg) or insertless plasmid pAc5.1/V5-His (mock) (1.0 μg). The luciferase activity of unmethylated pGL3-SNRPN in combination with pPacSp1 and pAc5.1/V5-His (mock) was normalized to 100 (data not shown). Relative luciferase activities (means + standard deviations) are given. The numbers 22, 30, 32, 34, 44, 45, and 52 correspond to R22A, R30A, D32A, Y34A, R44A, S45A, and Y52A, respectively.
FIG. 5
Residues within the methyl-CpG binding domain of MBD1 required for the methylated DNA binding, intranuclear localization, and transcriptional repression of methylated promoter. (A) Band shift of methylated DNA complexed with wild-type (wt) and mutant MBD1. Unmethylated (−) and _Sss_I-methylated (+) DNA fragments of the SNRPN promoter were incubated with MBD1 (residues 1 to 75) or GST. (B) Intranuclear localization of GFP-fused MBD1 (full-length and N-terminal deletion ΔN) and MBD1 (residues 1 to 89) with the above-mentioned point mutations. (C) _Hha_I- or _Sss_I-methylated pGL3-SNRPN (0.5 μg) was cotransfected with pPacSp1 (0.5 μg) and one of the MBD1-expressing plasmids [pAc5.1-MBD1v1, pAc5.1-MBD1v1ΔN, pAc5.1-wt. MBD1v1, pAc5.1-MBD1v1(R22A) to pAc5.1-MBD1v1(Y52A), pAc5.1-MBD1v3, pAc5.1-MBD1v3ΔN, pAc5.1-wt. MBD1v3, and pAc5.1-MBD1v3(R22A) to pAc5.1-MBD1v3(Y52A)] (1.0 μg) or insertless plasmid pAc5.1/V5-His (mock) (1.0 μg). The luciferase activity of unmethylated pGL3-SNRPN in combination with pPacSp1 and pAc5.1/V5-His (mock) was normalized to 100 (data not shown). Relative luciferase activities (means + standard deviations) are given. The numbers 22, 30, 32, 34, 44, 45, and 52 correspond to R22A, R30A, D32A, Y34A, R44A, S45A, and Y52A, respectively.
FIG. 6
TRD and DNA binding activity of CXXC3 in MBD1. (A) The reporter construct contains five copies of the GAL4 DNA binding site (5 × GAL) upstream of the SNRPN promoter. Effector constructs express the regions of MBD1 fused to the GAL4 DNA binding domain. CMV, cytomegalovirus promoter. (B) Transcriptional repression domain in MBD1 isoforms. Eleven GAL4 fusion proteins (termed GAL4-MBD1Δ1 to GAL4-MBD1Δ11) were constructed: Δ1 (amino acids 62 to 605 of MBD1v1), Δ2 (amino acids 62 to 549 of MBD1v3), Δ3 (amino acids 62 to 173 of MBD1v1), Δ4 (amino acids 62 to 327 of MBD1v3), Δ5 (amino acids 62 to 379 of MBD1v1), Δ6 (amino acids 380 to 605 of MBD1v1), Δ7 (amino acids 1 to 61 of MBD1v1), Δ8 (amino acids 361 to 586 of MBD1v2), Δ9 (amino acids 361 to 523 of MBD1v2), Δ10 (amino acids 361 to 459 of MBD1v2), and Δ11 (amino acids 460 to 523 of MBD1v2). +, shown; −, not shown. (C) Relative transcription levels under the expression of GAL4-MBD1Δ4, GAL4-MBD1Δ5, GAL4-MBD1Δ7, and GAL4-MBD1Δ11. GAL4-MBD1Δ11 specifically repressed transcription from the reporter. (D) Sodium dodecyl sulfate-polyacrylamide gel electrophoresis of GST-fused MBD1v1 and MBD1v3 stained with Coomassie blue: full-length (full) and with deletion of the MBD (amino acids 1 to 61) (ΔN). (E) Band shift analysis of methylated and unmethylated DNAs complexed with recombinant MBD1. Unmethylated (M−) and _Sss_I-methylated (M+) DNAs of the SNRPN promoter were incubated with one of the GST-MBD1 proteins. The CXXC3 of MBD1v1 has a DNA binding activity. Numbers to the right are molecular masses (in kilodaltons).
FIG. 6
TRD and DNA binding activity of CXXC3 in MBD1. (A) The reporter construct contains five copies of the GAL4 DNA binding site (5 × GAL) upstream of the SNRPN promoter. Effector constructs express the regions of MBD1 fused to the GAL4 DNA binding domain. CMV, cytomegalovirus promoter. (B) Transcriptional repression domain in MBD1 isoforms. Eleven GAL4 fusion proteins (termed GAL4-MBD1Δ1 to GAL4-MBD1Δ11) were constructed: Δ1 (amino acids 62 to 605 of MBD1v1), Δ2 (amino acids 62 to 549 of MBD1v3), Δ3 (amino acids 62 to 173 of MBD1v1), Δ4 (amino acids 62 to 327 of MBD1v3), Δ5 (amino acids 62 to 379 of MBD1v1), Δ6 (amino acids 380 to 605 of MBD1v1), Δ7 (amino acids 1 to 61 of MBD1v1), Δ8 (amino acids 361 to 586 of MBD1v2), Δ9 (amino acids 361 to 523 of MBD1v2), Δ10 (amino acids 361 to 459 of MBD1v2), and Δ11 (amino acids 460 to 523 of MBD1v2). +, shown; −, not shown. (C) Relative transcription levels under the expression of GAL4-MBD1Δ4, GAL4-MBD1Δ5, GAL4-MBD1Δ7, and GAL4-MBD1Δ11. GAL4-MBD1Δ11 specifically repressed transcription from the reporter. (D) Sodium dodecyl sulfate-polyacrylamide gel electrophoresis of GST-fused MBD1v1 and MBD1v3 stained with Coomassie blue: full-length (full) and with deletion of the MBD (amino acids 1 to 61) (ΔN). (E) Band shift analysis of methylated and unmethylated DNAs complexed with recombinant MBD1. Unmethylated (M−) and _Sss_I-methylated (M+) DNAs of the SNRPN promoter were incubated with one of the GST-MBD1 proteins. The CXXC3 of MBD1v1 has a DNA binding activity. Numbers to the right are molecular masses (in kilodaltons).
FIG. 6
TRD and DNA binding activity of CXXC3 in MBD1. (A) The reporter construct contains five copies of the GAL4 DNA binding site (5 × GAL) upstream of the SNRPN promoter. Effector constructs express the regions of MBD1 fused to the GAL4 DNA binding domain. CMV, cytomegalovirus promoter. (B) Transcriptional repression domain in MBD1 isoforms. Eleven GAL4 fusion proteins (termed GAL4-MBD1Δ1 to GAL4-MBD1Δ11) were constructed: Δ1 (amino acids 62 to 605 of MBD1v1), Δ2 (amino acids 62 to 549 of MBD1v3), Δ3 (amino acids 62 to 173 of MBD1v1), Δ4 (amino acids 62 to 327 of MBD1v3), Δ5 (amino acids 62 to 379 of MBD1v1), Δ6 (amino acids 380 to 605 of MBD1v1), Δ7 (amino acids 1 to 61 of MBD1v1), Δ8 (amino acids 361 to 586 of MBD1v2), Δ9 (amino acids 361 to 523 of MBD1v2), Δ10 (amino acids 361 to 459 of MBD1v2), and Δ11 (amino acids 460 to 523 of MBD1v2). +, shown; −, not shown. (C) Relative transcription levels under the expression of GAL4-MBD1Δ4, GAL4-MBD1Δ5, GAL4-MBD1Δ7, and GAL4-MBD1Δ11. GAL4-MBD1Δ11 specifically repressed transcription from the reporter. (D) Sodium dodecyl sulfate-polyacrylamide gel electrophoresis of GST-fused MBD1v1 and MBD1v3 stained with Coomassie blue: full-length (full) and with deletion of the MBD (amino acids 1 to 61) (ΔN). (E) Band shift analysis of methylated and unmethylated DNAs complexed with recombinant MBD1. Unmethylated (M−) and _Sss_I-methylated (M+) DNAs of the SNRPN promoter were incubated with one of the GST-MBD1 proteins. The CXXC3 of MBD1v1 has a DNA binding activity. Numbers to the right are molecular masses (in kilodaltons).
FIG. 6
TRD and DNA binding activity of CXXC3 in MBD1. (A) The reporter construct contains five copies of the GAL4 DNA binding site (5 × GAL) upstream of the SNRPN promoter. Effector constructs express the regions of MBD1 fused to the GAL4 DNA binding domain. CMV, cytomegalovirus promoter. (B) Transcriptional repression domain in MBD1 isoforms. Eleven GAL4 fusion proteins (termed GAL4-MBD1Δ1 to GAL4-MBD1Δ11) were constructed: Δ1 (amino acids 62 to 605 of MBD1v1), Δ2 (amino acids 62 to 549 of MBD1v3), Δ3 (amino acids 62 to 173 of MBD1v1), Δ4 (amino acids 62 to 327 of MBD1v3), Δ5 (amino acids 62 to 379 of MBD1v1), Δ6 (amino acids 380 to 605 of MBD1v1), Δ7 (amino acids 1 to 61 of MBD1v1), Δ8 (amino acids 361 to 586 of MBD1v2), Δ9 (amino acids 361 to 523 of MBD1v2), Δ10 (amino acids 361 to 459 of MBD1v2), and Δ11 (amino acids 460 to 523 of MBD1v2). +, shown; −, not shown. (C) Relative transcription levels under the expression of GAL4-MBD1Δ4, GAL4-MBD1Δ5, GAL4-MBD1Δ7, and GAL4-MBD1Δ11. GAL4-MBD1Δ11 specifically repressed transcription from the reporter. (D) Sodium dodecyl sulfate-polyacrylamide gel electrophoresis of GST-fused MBD1v1 and MBD1v3 stained with Coomassie blue: full-length (full) and with deletion of the MBD (amino acids 1 to 61) (ΔN). (E) Band shift analysis of methylated and unmethylated DNAs complexed with recombinant MBD1. Unmethylated (M−) and _Sss_I-methylated (M+) DNAs of the SNRPN promoter were incubated with one of the GST-MBD1 proteins. The CXXC3 of MBD1v1 has a DNA binding activity. Numbers to the right are molecular masses (in kilodaltons).
FIG. 6
TRD and DNA binding activity of CXXC3 in MBD1. (A) The reporter construct contains five copies of the GAL4 DNA binding site (5 × GAL) upstream of the SNRPN promoter. Effector constructs express the regions of MBD1 fused to the GAL4 DNA binding domain. CMV, cytomegalovirus promoter. (B) Transcriptional repression domain in MBD1 isoforms. Eleven GAL4 fusion proteins (termed GAL4-MBD1Δ1 to GAL4-MBD1Δ11) were constructed: Δ1 (amino acids 62 to 605 of MBD1v1), Δ2 (amino acids 62 to 549 of MBD1v3), Δ3 (amino acids 62 to 173 of MBD1v1), Δ4 (amino acids 62 to 327 of MBD1v3), Δ5 (amino acids 62 to 379 of MBD1v1), Δ6 (amino acids 380 to 605 of MBD1v1), Δ7 (amino acids 1 to 61 of MBD1v1), Δ8 (amino acids 361 to 586 of MBD1v2), Δ9 (amino acids 361 to 523 of MBD1v2), Δ10 (amino acids 361 to 459 of MBD1v2), and Δ11 (amino acids 460 to 523 of MBD1v2). +, shown; −, not shown. (C) Relative transcription levels under the expression of GAL4-MBD1Δ4, GAL4-MBD1Δ5, GAL4-MBD1Δ7, and GAL4-MBD1Δ11. GAL4-MBD1Δ11 specifically repressed transcription from the reporter. (D) Sodium dodecyl sulfate-polyacrylamide gel electrophoresis of GST-fused MBD1v1 and MBD1v3 stained with Coomassie blue: full-length (full) and with deletion of the MBD (amino acids 1 to 61) (ΔN). (E) Band shift analysis of methylated and unmethylated DNAs complexed with recombinant MBD1. Unmethylated (M−) and _Sss_I-methylated (M+) DNAs of the SNRPN promoter were incubated with one of the GST-MBD1 proteins. The CXXC3 of MBD1v1 has a DNA binding activity. Numbers to the right are molecular masses (in kilodaltons).
Similar articles
- Regulation of transcription and chromatin by methyl-CpG binding protein MBD1.
Nakao M, Matsui S, Yamamoto S, Okumura K, Shirakawa M, Fujita N. Nakao M, et al. Brain Dev. 2001 Dec;23 Suppl 1:S174-6. doi: 10.1016/s0387-7604(01)00348-5. Brain Dev. 2001. PMID: 11738867 Review. - Methylation-mediated transcriptional silencing in euchromatin by methyl-CpG binding protein MBD1 isoforms.
Fujita N, Takebayashi S, Okumura K, Kudo S, Chiba T, Saya H, Nakao M. Fujita N, et al. Mol Cell Biol. 1999 Sep;19(9):6415-26. doi: 10.1128/MCB.19.9.6415. Mol Cell Biol. 1999. PMID: 10454587 Free PMC article. - Mbd1 is recruited to both methylated and nonmethylated CpGs via distinct DNA binding domains.
Jørgensen HF, Ben-Porath I, Bird AP. Jørgensen HF, et al. Mol Cell Biol. 2004 Apr;24(8):3387-95. doi: 10.1128/MCB.24.8.3387-3395.2004. Mol Cell Biol. 2004. PMID: 15060159 Free PMC article. - Methyl-CpG binding domain 1 (MBD1) interacts with the Suv39h1-HP1 heterochromatic complex for DNA methylation-based transcriptional repression.
Fujita N, Watanabe S, Ichimura T, Tsuruzoe S, Shinkai Y, Tachibana M, Chiba T, Nakao M. Fujita N, et al. J Biol Chem. 2003 Jun 27;278(26):24132-8. doi: 10.1074/jbc.M302283200. Epub 2003 Apr 23. J Biol Chem. 2003. PMID: 12711603 - An epigenetic regulator: methyl-CpG-binding domain protein 1 (MBD1).
Li L, Chen BF, Chan WY. Li L, et al. Int J Mol Sci. 2015 Mar 5;16(3):5125-40. doi: 10.3390/ijms16035125. Int J Mol Sci. 2015. PMID: 25751725 Free PMC article. Review.
Cited by
- FAD-dependent lysine-specific demethylase-1 regulates cellular energy expenditure.
Hino S, Sakamoto A, Nagaoka K, Anan K, Wang Y, Mimasu S, Umehara T, Yokoyama S, Kosai K, Nakao M. Hino S, et al. Nat Commun. 2012 Mar 27;3:758. doi: 10.1038/ncomms1755. Nat Commun. 2012. PMID: 22453831 Free PMC article. - Pyrrole-imidazole hairpin polyamides with high affinity at 5'-CGCG-3' DNA sequence; influence of cytosine methylation on binding.
Minoshima M, Bando T, Sasaki S, Fujimoto J, Sugiyama H. Minoshima M, et al. Nucleic Acids Res. 2008 May;36(9):2889-94. doi: 10.1093/nar/gkn116. Epub 2008 Apr 1. Nucleic Acids Res. 2008. PMID: 18385159 Free PMC article. - High mobility group protein HMGA1 inhibits retinoblastoma protein-mediated cellular G0 arrest.
Ueda Y, Watanabe S, Tei S, Saitoh N, Kuratsu J, Nakao M. Ueda Y, et al. Cancer Sci. 2007 Dec;98(12):1893-901. doi: 10.1111/j.1349-7006.2007.00608.x. Epub 2007 Sep 17. Cancer Sci. 2007. PMID: 17877762 Free PMC article. - Mice lacking methyl-CpG binding protein 1 have deficits in adult neurogenesis and hippocampal function.
Zhao X, Ueba T, Christie BR, Barkho B, McConnell MJ, Nakashima K, Lein ES, Eadie BD, Willhoite AR, Muotri AR, Summers RG, Chun J, Lee KF, Gage FH. Zhao X, et al. Proc Natl Acad Sci U S A. 2003 May 27;100(11):6777-82. doi: 10.1073/pnas.1131928100. Epub 2003 May 14. Proc Natl Acad Sci U S A. 2003. PMID: 12748381 Free PMC article. - Methyl-CpG-binding protein 2 represses LINE-1 expression and retrotransposition but not Alu transcription.
Yu F, Zingler N, Schumann G, Strätling WH. Yu F, et al. Nucleic Acids Res. 2001 Nov 1;29(21):4493-501. doi: 10.1093/nar/29.21.4493. Nucleic Acids Res. 2001. PMID: 11691937 Free PMC article.
References
- Ballabio A, Willard H F. Mammalian X-chromosome inactivation and the XIST gene. Curr Opin Genet Dev. 1992;2:439–447. - PubMed
- Bartolomei M S, Tilghman S M. Genomic imprinting in mammals. Annu Rev Genet. 1997;31:493–525. - PubMed
- Bestor T H, Verdine G L. DNA methyltransferases. Curr Opin Cell Biol. 1994;6:380–389. - PubMed
- Bird A P, Wolffe A P. Methylation-induced repression-belts, braces, and chromatin. Cell. 1999;99:451–454. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials