Analysis of MinC reveals two independent domains involved in interaction with MinD and FtsZ - PubMed (original) (raw)
Comparative Study
Analysis of MinC reveals two independent domains involved in interaction with MinD and FtsZ
Z Hu et al. J Bacteriol. 2000 Jul.
Abstract
In Escherichia coli FtsZ assembles into a Z ring at midcell while assembly at polar sites is prevented by the min system. MinC, a component of this system, is an inhibitor of FtsZ assembly that is positioned within the cell by interaction with MinDE. In this study we found that MinC consists of two functional domains connected by a short linker. When fused to MalE the N-terminal domain is able to inhibit cell division and prevent FtsZ assembly in vitro. The C-terminal domain interacts with MinD, and expression in wild-type cells as a MalE fusion disrupts min function, resulting in a minicell phenotype. We also find that MinC is an oligomer, probably a dimer. Although the C-terminal domain is clearly sufficient for oligomerization, the N-terminal domain also promotes oligomerization. These results demonstrate that MinC consists of two independently functioning domains: an N-terminal domain capable of inhibiting FtsZ assembly and a C-terminal domain responsible for localization of MinC through interaction with MinD. The fusion of these two independent domains is required to achieve topological regulation of Z ring assembly.
Figures
FIG. 1
Alignment of MinC sequences. The sequences of MinC from various bacteria were aligned using MegAlign (DNA Star) and the Clustal Method. Identical amino acids in three or more sequences are boxed. The MinC sequences (with GenBank accession numbers in parentheses) are, from the top, E. coli (AAB59061.1), S. enterica serovar Typhimurium, V. cholerae, Bacillus subtilis (AAA22400.1), and T. maritima (AAD36124.1). The asterisk indicates that the B. subtilis MinC is truncated and the last 15 residues are not shown.
FIG. 2
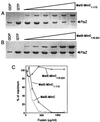
Gel filtration chromatography of MalE-MinC and MalE-MinC19. Affinity-purified MalE-MinC was analyzed on a Superose-6 gel filtration column equilibrated with polymerization buffer. Fractions obtained from the elution were analyzed by SDS-PAGE (fraction number indicated at the top). (A and B) Lane S contains molecular weight markers (from the top, phosphorylase b, 97.4K; serum albumin, 66K; ovalbumin, 45K; and carbonic anhydrase, 29K). (A) A 1-ml sample of MalE-MinC (12.5 μM) was applied to the column; (B) a 1-ml sample of MalE-MinC19 (12 .5 μM) was applied to the column. (C) A standard curve for estimating the size of MalE-MinC was obtained by running the following molecular weight standards: apoferritin (400K), β-amylase (200K), alcohol dehydrogenase (150K), bovine serum albumin (66K), and carbonic anhydrase (29K).
FIG. 3
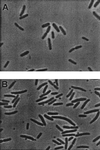
Expression of the C-terminal domain of MinC induces minicell formation in wild-type cells. JS219 containing pJC90 (malE) (A) or pZH111 (_malE-minC_116-231) (B) was diluted from an overnight culture and grown for several hours. Arabinose (0.005%) was added, and samples were taken for photography 2 h later.
FIG. 4
Expression of the N-terminal domain of MinC induces filamentation. JS964 (Δ_min_) containing various MalE fusions was photographed 90 to 120 min after adding arabinose (0.005%) to exponentially growing cultures. The plasmids and fusions used were as follows: (A) pJC90 (malE), (B) pZH111 (_malE-minC_1–115), (C) pZH112 (_malE-minC_116–231), and (D) pZH101 (malE-minC).
FIG. 5
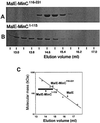
The N-terminal domain of MinC is sufficient to prevent FtsZ polymerization. Affinity-purified MalE-MinC1–115 and MalE-MinC116–231 were tested for their effect on FtsZ polymerization utilizing a sedimentation assay. FtsZ at 200 μg/ml was incubated in polymerization buffer (50 mM morpholineethanesulfonic acid [pH 6.5], 50 mM KCl, 1 mM MgCl2) with increasing concentrations of the MalE fusions. The reactions were initiated by the addition of GTP at 1 mM. After a 5-min incubation at room temperature the samples were centrifuged at 80K rpm for 15 min in a Beckman TLA 100.2 rotor. The pellets were resuspended in SDS sample buffer and analyzed by SDS-PAGE. (A and B) Lanes GDP contain a control with GDP added, and lanes GTP contain GTP but no fusion protein. The final concentration of fusion protein added (in micrograms per milliliter) was 50, 100, 200, 400, 800, and 1,200 in lanes 3 to 8, respectively. (C) The amount of FtsZ in the pellet was plotted as a percentage of the control lacking the fusion protein. The data for MalE-MinC was taken from reference .
FIG. 6
The C-terminal and N-terminal domains of MinC promote oligomerization. MalE-MinC1–115 and MalE-MinC116–231 were analyzed by gel filtration chromatography as described in the legend to Fig. 2, except that smaller fractions were collected. (A and B) Fractions from the elution were analyzed by SDS-PAGE. (C) The same standard curve shown in Fig. 2 was used to estimate the size of the fusions.
FIG. 7
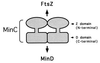
Model of MinC. In this model MinC is depicted as a dimer although it is possible that it forms larger oligomers. The N-terminal domain (Z domain) interacts with FtsZ to prevent polymerization. The C-terminal domain (D domain) is responsible for interaction with MinD resulting in placement of MinC at the membrane. It is not clear if dimerization plays a role in these interactions. The C-terminal domain is clearly sufficient for dimerization, although in vitro results show that the N-terminal domain may also contribute to dimerization. The N-terminal domain also promotes the formation of oligomers larger than dimers. This activity is partially suppressed in the full-length MinC.
Similar articles
- MinC N- and C-Domain Interactions Modulate FtsZ Assembly, Division Site Selection, and MinD-Dependent Oscillation in Escherichia coli.
LaBreck CJ, Conti J, Viola MG, Camberg JL. LaBreck CJ, et al. J Bacteriol. 2019 Jan 28;201(4):e00374-18. doi: 10.1128/JB.00374-18. Print 2019 Feb 15. J Bacteriol. 2019. PMID: 30455283 Free PMC article. - The conserved C-terminal tail of FtsZ is required for the septal localization and division inhibitory activity of MinC(C)/MinD.
Shen B, Lutkenhaus J. Shen B, et al. Mol Microbiol. 2009 Apr;72(2):410-24. doi: 10.1111/j.1365-2958.2009.06651.x. Mol Microbiol. 2009. PMID: 19415799 Free PMC article. - Targeting of (D)MinC/MinD and (D)MinC/DicB complexes to septal rings in Escherichia coli suggests a multistep mechanism for MinC-mediated destruction of nascent FtsZ rings.
Johnson JE, Lackner LL, de Boer PA. Johnson JE, et al. J Bacteriol. 2002 Jun;184(11):2951-62. doi: 10.1128/JB.184.11.2951-2962.2002. J Bacteriol. 2002. PMID: 12003935 Free PMC article. - The MinC component of the division site selection system in Escherichia coli interacts with FtsZ to prevent polymerization.
Hu Z, Mukherjee A, Pichoff S, Lutkenhaus J. Hu Z, et al. Proc Natl Acad Sci U S A. 1999 Dec 21;96(26):14819-24. doi: 10.1073/pnas.96.26.14819. Proc Natl Acad Sci U S A. 1999. PMID: 10611296 Free PMC article. - The keepers of the ring: regulators of FtsZ assembly.
Ortiz C, Natale P, Cueto L, Vicente M. Ortiz C, et al. FEMS Microbiol Rev. 2016 Jan;40(1):57-67. doi: 10.1093/femsre/fuv040. Epub 2015 Sep 15. FEMS Microbiol Rev. 2016. PMID: 26377318 Review.
Cited by
- MinC protein shortens FtsZ protofilaments by preferentially interacting with GDP-bound subunits.
Hernández-Rocamora VM, García-Montañés C, Reija B, Monterroso B, Margolin W, Alfonso C, Zorrilla S, Rivas G. Hernández-Rocamora VM, et al. J Biol Chem. 2013 Aug 23;288(34):24625-35. doi: 10.1074/jbc.M113.483222. Epub 2013 Jul 12. J Biol Chem. 2013. PMID: 23853099 Free PMC article. - Cytokinesis in bacteria.
Errington J, Daniel RA, Scheffers DJ. Errington J, et al. Microbiol Mol Biol Rev. 2003 Mar;67(1):52-65, table of contents. doi: 10.1128/MMBR.67.1.52-65.2003. Microbiol Mol Biol Rev. 2003. PMID: 12626683 Free PMC article. Review. - Dimerization or oligomerization of the actin-like FtsA protein enhances the integrity of the cytokinetic Z ring.
Shiomi D, Margolin W. Shiomi D, et al. Mol Microbiol. 2007 Dec;66(6):1396-415. doi: 10.1111/j.1365-2958.2007.05998.x. Epub 2007 Nov 6. Mol Microbiol. 2007. PMID: 17986188 Free PMC article. - The dimerization function of MinC resides in a structurally autonomous C-terminal domain.
Szeto TH, Rowland SL, King GF. Szeto TH, et al. J Bacteriol. 2001 Nov;183(22):6684-7. doi: 10.1128/JB.183.22.6684-6687.2001. J Bacteriol. 2001. PMID: 11673440 Free PMC article. - The double par locus of virulence factor pB171: DNA segregation is correlated with oscillation of ParA.
Ebersbach G, Gerdes K. Ebersbach G, et al. Proc Natl Acad Sci U S A. 2001 Dec 18;98(26):15078-83. doi: 10.1073/pnas.261569598. Proc Natl Acad Sci U S A. 2001. PMID: 11752455 Free PMC article.
References
- Bartel P L, Chien C-T, Sternglanz R, Fields S. Elimination of false positives that arise in using the two-hybrid system. BioTechniques. 1993;14:920–924. - PubMed
- Bi E, Lutkenhaus J. FtsZ ring structure associated with division in Escherichia coli. Nature. 1991;354:161–164. - PubMed
- de Boer P A J, Crossley R E, Rothfield L I. A division inhibitor and a topological specificity factor coded for by the minicell locus determine the proper placement of the division site in Escherichia coli. Cell. 1989;56:641–649. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases