The ancestry of Brazilian mtDNA lineages - PubMed (original) (raw)
The ancestry of Brazilian mtDNA lineages
J Alves-Silva et al. Am J Hum Genet. 2000 Aug.
Erratum in
- Am J Hum Genet 2000 Sep;67(3):775
Abstract
We have analyzed 247 Brazilian mtDNAs for hypervariable segment (HVS)-I and selected restriction fragment-length-polymorphism sites, to assess their ancestry in different continents. The total sample showed nearly equal amounts of Native American, African, and European matrilineal genetic contribution but with regional differences within Brazil. The mtDNA pool of present-day Brazilians clearly reflects the imprints of the early Portuguese colonization process (involving directional mating), as well as the recent immigrant waves (from Europe) of the last century. The subset of 99 mtDNAs from the southeastern region encompasses nearly all mtDNA haplogroups observed in the total Brazilian sample; for this regional subset, HVS-II was analyzed, providing, in particular, some novel details of the African mtDNA phylogeny.
Figures
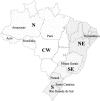
Figure 1
Five major geographic regions of Brazil: N = northern, NE = northeastern, SE = southeastern, S = southern, and CW = central west. Brazilian states from which the mtDNA lineages of the present study have mainly been sampled are indicated by name.
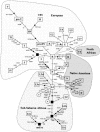
Figure 2
Classification tree highlighting selected diagnostic sites and positions for haplogroups present in the Brazilian sample (see tables 1 andtable 6table 6). Each square represents the root node of the respective haplogroup, with the acronym inscribed; two central/eastern-African haplogroups, represented by circles, are only partially characterized (T. Kisivild, personal communication). “CRS” indicates the revised reference sequence (Andrews et al. 1999). Numbers along links refer to RFLP sites (with arrows pointing to presence of sites) or transitions, unless a single-letter suffix indicates a transversion. Note that some diagnostic sites and positions, especially in the control region, have undergone recurrent mutations. The root of the tree, labeled “mtEve,” is inferred by employing the Neanderthal HVS-I and HVS-II sequences (Krings et al. 1997, 1999) and the coding-region sequences of bonobo and common chimpanzee (Horai et al. 1995) as outgroups; this corroborates the rooting of the Vigilant (1990) tree.
Similar articles
- Multiple geographic sources of region V 9-bp deletion haplotypes in Brazilians.
Alves-Silva J, Santos MS, Pena SD, Prado VF. Alves-Silva J, et al. Hum Biol. 1999 Apr;71(2):245-59. Hum Biol. 1999. PMID: 10222646 - mtDNA structure: the women who formed the Brazilian Northeast.
Schaan AP, Costa L, Santos D, Modesto A, Amador M, Lopes C, Rabenhorst SH, Montenegro R, Souza BDA, Lopes T, Yoshioka FK, Pinto G, Silbiger V, Ribeiro-Dos-Santos Â. Schaan AP, et al. BMC Evol Biol. 2017 Aug 9;17(1):185. doi: 10.1186/s12862-017-1027-7. BMC Evol Biol. 2017. PMID: 28793858 Free PMC article. - mtDNA haplogroup X: An ancient link between Europe/Western Asia and North America?
Brown MD, Hosseini SH, Torroni A, Bandelt HJ, Allen JC, Schurr TG, Scozzari R, Cruciani F, Wallace DC. Brown MD, et al. Am J Hum Genet. 1998 Dec;63(6):1852-61. doi: 10.1086/302155. Am J Hum Genet. 1998. PMID: 9837837 Free PMC article. - Autosomal, mtDNA, and Y-chromosome diversity in Amerinds: pre- and post-Columbian patterns of gene flow in South America.
Mesa NR, Mondragón MC, Soto ID, Parra MV, Duque C, Ortíz-Barrientos D, García LF, Velez ID, Bravo ML, Múnera JG, Bedoya G, Bortolini MC, Ruiz-Linares A. Mesa NR, et al. Am J Hum Genet. 2000 Nov;67(5):1277-86. doi: 10.1016/S0002-9297(07)62955-3. Epub 2000 Oct 13. Am J Hum Genet. 2000. PMID: 11032789 Free PMC article. - mtDNA affinities of the peoples of North-Central Mexico.
Green LD, Derr JN, Knight A. Green LD, et al. Am J Hum Genet. 2000 Mar;66(3):989-98. doi: 10.1086/302801. Am J Hum Genet. 2000. PMID: 10712213 Free PMC article.
Cited by
- A pilot study of mitochondrial genomic ancestry in admixed Brazilian patients with type 1 diabetes.
Ferreira LL, Gonçalves ABR, Adiala IJB, Loiola S, Dias A, Azulay RS, Silva DA, Gomes MB. Ferreira LL, et al. Diabetol Metab Syndr. 2024 Jun 15;16(1):130. doi: 10.1186/s13098-024-01342-8. Diabetol Metab Syndr. 2024. PMID: 38879575 Free PMC article. - Maternal ancestry and lineages diversity of the Santander population from Colombia.
Castillo A, Rondón F, Mantilla G, Gusmão L, Simão F. Castillo A, et al. Forensic Sci Res. 2023 Sep 21;8(3):241-248. doi: 10.1093/fsr/owad032. eCollection 2023 Sep. Forensic Sci Res. 2023. PMID: 38221971 Free PMC article. - Human N-acetyltransferase 2 (NAT2) gene variability in Brazilian populations from different geographical areas.
Lopes MQP, Teixeira RLF, Cabello PH, Nery JAC, Sales AM, Nahn J R EP, Moreira MV, Stahlke EVR, Possuelo LG, Rossetti MLR, Rabahi MF, Silva LFM, Leme PA, Woods WJ, Nobre ML, de Oliveira MLW, Narahashi K, Cavalcanti M, Suffys PN, Boukouvala S, Gallo MEN, Santos AR. Lopes MQP, et al. Front Pharmacol. 2023 Nov 15;14:1278720. doi: 10.3389/fphar.2023.1278720. eCollection 2023. Front Pharmacol. 2023. PMID: 38035025 Free PMC article. - High Mitochondrial Haplotype Diversity Found in Three Pre-Hispanic Groups from Colombia.
Uricoechea Patiño D, Collins A, García OJR, Santos Vecino G, Cuenca JVR, Bernal JE, Benavides Benítez E, Vergara Muñoz S, Briceño Balcázar I. Uricoechea Patiño D, et al. Genes (Basel). 2023 Sep 23;14(10):1853. doi: 10.3390/genes14101853. Genes (Basel). 2023. PMID: 37895202 Free PMC article. - A systematic comparison of human mitochondrial genome assembly tools.
Mahar NS, Satyam R, Sundar D, Gupta I. Mahar NS, et al. BMC Bioinformatics. 2023 Sep 13;24(1):341. doi: 10.1186/s12859-023-05445-3. BMC Bioinformatics. 2023. PMID: 37704952 Free PMC article.
References
Electronic-Database Information
- GenBank Overview, http://www.ncbi.nlm.nih.gov/Genbank/GenbankOverview.html (for HVS-I [accession numbers AF243627–AF243796] and HVS-II [accession numbers AF243539–AF243626])
- Instituto Brasileiro de Geografia Estatística, http://www.ibge.gov.br/
References
- Alves-Silva J, Guimarães PE, Rocha J, Pena SD, Prado VF (1999) Identification in Portugal and Brazil of a mtDNA lineage containing a 9-bp triplication of the intergenic COII/tRNALys region. Hum Hered 49:56–58 - PubMed
- Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N (1999) Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet 23:147 - PubMed
- Arpini-Sampaio Z, Costa MC, Melo AA, Carvalho MF, Deus MS, Simões AL (1999) Genetic polymorphisms and ethnic admixture in African-derived black communities of northeastern Brazil. Hum Biol 71:69–85 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources