Sister chromatid gene conversion is a prominent double-strand break repair pathway in mammalian cells - PubMed (original) (raw)
Sister chromatid gene conversion is a prominent double-strand break repair pathway in mammalian cells
R D Johnson et al. EMBO J. 2000.
Abstract
In mammalian cells, repair of DNA double-strand breaks (DSBs) occurs by both homologous and non-homologous mechanisms. By definition, homologous recombination requires a template with sufficient sequence identity to the damaged molecule in order to direct repair. We now show that the sister chromatid acts as a repair template in a substantial proportion of DSB repair events. The outcome of sister chromatid repair is primarily gene conversion unassociated with reciprocal exchange. This contrasts with expectations from the classical DSB repair model originally proposed for yeast meiotic recombination, but is consistent with models in which recombination is coupled intimately with replication. These results may explain why cytologically observable sister chromatid exchanges are induced only weakly by DNA-damaging agents that cause strand breaks, since most homologous repair events would not be observed. A preference for non-crossover events between sister chromatids suggests that crossovers, although genetically silent, may be disfavored for other reasons. Possibly, a general bias against crossing over in mitotic cells exists to reduce the potential for genome alterations when other homologous repair templates are utilized.
Figures
Fig. 1. Recombination reporter substrate SCneo. (A) Schematic representation of SCneo and predicted neo+ and _neo_– DSB repair products after I-_Sce_I expression. Shaded boxes represent the 0.7 kb neo gene repeats and the open box the promoter of the S2neo gene. The small black box is the 18 bp I-_Sce_I site. During a short tract gene conversion (STGC), the I-_Sce_I site is replaced by the _Nco_I site, but SCneo is not otherwise altered. Both long tract gene conversion and unequal sister chromatid exchange (LTGC/SCE) results in expansion of the SCneo locus. The expansion typically results in the middle neo repeat containing _Nco_I and _Sal_I sites. Other LTGC products are also possible, depending on the length of the conversion tract. In a non-homologous end joining (NHEJ) event, the I-_Sce_I site is disrupted, as these events usually involve small deletions or insertions. The 0.7 kb product can arise either from a homologous deletion (HD) or as the reciprocal contraction product of an unequal SCE. HD events arise from annealing of single strands at the two neo repeats, such that one repeat and the intervening hyg gene is deleted. The relevant restriction enzyme sites are shown: H, _Hin_dIII; N, _Nco_I; S, _Sal_I; B, _Bam_HI; X, _Xho_I. (B) Predicted products from two distinct mechanisms of unequal sister chromatid recombination. In an LTGC, the donor of information will remain unaltered during conversion of sequences downstream of the I-_Sce_I site. In an SCE, expansion of one chromatid to a triplication will be associated with contraction on the other chromatid.
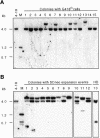
Fig. 2. Sister chromatid recombination within the SCneo locus is not associated with reciprocal exchange. After I-_Sce_I expression in cell lines containing the SCneo substrate, single cells were grown in non-selective medium and then replica plated to test for resistance to G418. Southern blot analysis was performed on clones containing G418R cells using _Xho_I and _Hin_dIII digestion of genomic DNA. The probe was a 1.2 kb _Bam_HI–_Xho_I DNA fragment containing the complete neo sequence (Figure 1A). (A) Parental V79 (4-18) cells and clones in which G418R cells were identified. STGC events, lanes 1–4, 6, 8, 9, 12, 14 and 15; LTGC events, lanes 5, 7, 10, 11 and 13. (B) Parental V79 (4-18) cells and clones in which cells were identified to have undergone an LTGC event (lanes 1–12) or an HD event (lane 13). In both (A) and (B), each of the expansion products is presumed to be derived from an LTGC event, as none of the clones contains an associated 0.7 kb reciprocal SCE product. M, marker derived from V79 (4-18) genomic DNA digested with _Xho_I, _Bam_HI and _Hin_dIII.
Fig. 3. Segregation of donor and recipient sequences after sister chromatid recombination. A clone containing 7.3 and 4.0 kb _Xho_I–_Hin_dIII SCneo fragments was subcloned. Each subclone contains either the 4.0 or the 7.3 kb _Xho_I–_Hin_dIII fragment as expected for segregation of sister chromatids to daughter cells. Presumably the 4.0 kb fragment served as the donor of information during DSB repair by LTGC to generate the 7.3 kb fragment on the recipient chromatid. Genomic DNA from each subclone was digested with _Xho_I and _Hin_dIII and subjected to Southern blot analysis using a neo gene probe.
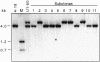
Fig. 4. Structure of sister chromatid recombination products. To derive the structures, genomic DNA from each LTGC clone was digested with the indicated restriction enzymes and subjected to Southern blot analysis. The total number of events obtained for each class is indicated. H, _Hin_dIII; N, _Nco_I; S, _Sal_I; B, _Bam_HI; Bst, _Bst_XI; Sac, _Sac_I; Spe, _Spe_I; X, _Xho_I, (X,H), either a _Xho_I or _Hin_dIII site.
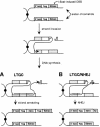
Fig. 5. Model for the generation of sister chromatid gene conversion events. LTGC and LTGC/NHEJ events can be initiated by strand invasion of the broken S2neo gene into the 3′ neo gene on the sister chromatid, with repair synthesis primed by the invading end. (A) In an LTGC event, repair synthesis continues into the downstream S2neo gene and is followed by homologous annealing of the newly synthesized strand to the end of the broken chromatid. (B) In an LTGC/NHEJ event, repair synthesis terminates downstream of the neo+ gene and is followed by NHEJ of the newly synthesized strand to the end of the broken chromatid. See text for details.
Similar articles
- Double-strand-break-induced homologous recombination in mammalian cells.
Johnson RD, Jasin M. Johnson RD, et al. Biochem Soc Trans. 2001 May;29(Pt 2):196-201. doi: 10.1042/0300-5127:0290196. Biochem Soc Trans. 2001. PMID: 11356153 Review. - Mouse RAD54 affects DNA double-strand break repair and sister chromatid exchange.
Dronkert ML, Beverloo HB, Johnson RD, Hoeijmakers JH, Jasin M, Kanaar R. Dronkert ML, et al. Mol Cell Biol. 2000 May;20(9):3147-56. doi: 10.1128/MCB.20.9.3147-3156.2000. Mol Cell Biol. 2000. PMID: 10757799 Free PMC article. - Analysis of repair of replication-born double-strand breaks by sister chromatid recombination in yeast.
Gómez-González B, Ortega P, Aguilera A. Gómez-González B, et al. Methods Enzymol. 2021;661:121-138. doi: 10.1016/bs.mie.2021.08.010. Epub 2021 Sep 21. Methods Enzymol. 2021. PMID: 34776209 - Pathways for mitotic homologous recombination in mammalian cells.
Helleday T. Helleday T. Mutat Res. 2003 Nov 27;532(1-2):103-15. doi: 10.1016/j.mrfmmm.2003.08.013. Mutat Res. 2003. PMID: 14643432 Review.
Cited by
- Genetic variation in recalcitrant repetitive regions of the Drosophila melanogaster genome.
Shukla HG, Chakraborty M, Emerson JJ. Shukla HG, et al. bioRxiv [Preprint]. 2024 Jun 12:2024.06.11.598575. doi: 10.1101/2024.06.11.598575. bioRxiv. 2024. PMID: 38915508 Free PMC article. Preprint. - Cell-type-specific consequences of mosaic structural variants in hematopoietic stem and progenitor cells.
Grimes K, Jeong H, Amoah A, Xu N, Niemann J, Raeder B, Hasenfeld P, Stober C, Rausch T, Benito E, Jann JC, Nowak D, Emini R, Hoenicka M, Liebold A, Ho A, Shuai S, Geiger H, Sanders AD, Korbel JO. Grimes K, et al. Nat Genet. 2024 Jun;56(6):1134-1146. doi: 10.1038/s41588-024-01754-2. Epub 2024 May 28. Nat Genet. 2024. PMID: 38806714 Free PMC article. - The impact of developmental stage, tissue type, and sex on DNA double-strand break repair in Drosophila melanogaster.
Graham EL, Fernandez J, Gandhi S, Choudhry I, Kellam N, LaRocque JR. Graham EL, et al. PLoS Genet. 2024 Apr 29;20(4):e1011250. doi: 10.1371/journal.pgen.1011250. eCollection 2024 Apr. PLoS Genet. 2024. PMID: 38683763 Free PMC article. - Developmental progression of DNA double-strand break repair deciphered by a single-allele resolution mutation classifier.
Li Z, You L, Hermann A, Bier E. Li Z, et al. Nat Commun. 2024 Mar 23;15(1):2629. doi: 10.1038/s41467-024-46479-2. Nat Commun. 2024. PMID: 38521791 Free PMC article. - Natural Biopolymer-Based Delivery of CRISPR/Cas9 for Cancer Treatment.
Lin M, Wang X. Lin M, et al. Pharmaceutics. 2023 Dec 30;16(1):62. doi: 10.3390/pharmaceutics16010062. Pharmaceutics. 2023. PMID: 38258073 Free PMC article. Review.
References
- Carrano A.V., Thompson,L.H., Lindl,P.A. and Minkler,J.L. (1978) Sister chromatid exchange as an indicator of mutagenesis. Nature, 271, 551–553. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources