SEL-8, a nuclear protein required for LIN-12 and GLP-1 signaling in Caenorhabditis elegans - PubMed (original) (raw)
Comparative Study
SEL-8, a nuclear protein required for LIN-12 and GLP-1 signaling in Caenorhabditis elegans
T G Doyle et al. Proc Natl Acad Sci U S A. 2000.
Abstract
LIN-12 and GLP-1 are members of the LIN-12/Notch family of receptors that mediate cell-cell interactions during development. The sel-8 gene had been identified previously in a screen for suppressors of a mutation that constitutively activates LIN-12. Here, we report that sel-8 is essential for lin-12- and glp-1-mediated signaling, and that SEL-8 is a glutamine-rich nuclear protein. We postulate that SEL-8 serves as a transcriptional coactivator or as an assembly factor for transcription complexes that contain the LIN-12 or GLP-1 intracellular domains.
Figures
Figure 1
Sequence analysis. (A) Schematic representation of transcripts from the sel-8 gene. RACE analysis revealed two potential transcripts, sel-8A and sel-8B. The sel-8(sa54) C-to-T mutation is indicated (nucleotide 17,295 of cosmid C32A3, GenBank accession no. Z48241). Introns are not drawn to scale. (B) Schematic representation of the SEL-8 protein. The “SEL-8 motif” that is found in W09C5.1 and its orthologs, the PEST sequence, and the two glutamine-rich regions are indicated and described further in the text. (C) Amino acid sequence of SEL-8. The sequence based on the sel-8A RACE product is shown. The PEST sequence is underlined with a solid line, and the motif shared with W09C5.1 is underlined with a dashed line. Glutamine (Q) residues are indicated in bold type. The boxed glutamine corresponds to codon 387 of sel-8A, which is changed to “stop” in sel-8(sa54). (D) Sequence motif found in SEL-8 and W09C5.1 (GenBank accession no. T26298) and its apparent yeast (YER126c, GenBank accession no. S43218) and Drosophila (DMIP259, GenBank accession no. AAF52940) orthologs. Reverse-contrast lettering indicates amino acids that are identical in SEL-8 and at least two of the other proteins.
Figure 2
SEL-8∷GFP localization. Epifluorescence micrographs of living arIs50 individuals show nuclear localization of SEL-8∷GFP. (A) Early embryo. (B) L2 hermaphrodite, somatic gonad. The arrow indicates Z1.ppp (Z4.aaa is in a different focal plane). The other fluorescent nuclei visualized in this focal plane are in the ventral cord.
Figure 3
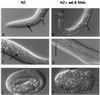
RNA-mediated interference. Nomarski photomicrographs of the progeny of untreated or mock-injected N2 hermaphrodites (Left) and the progeny of hermaphrodites that had been injected with sel-8 dsRNA (Right). (A and B) Head region, L1 larvae. Black arrow indicates the anterior bulb of the pharynx (missing in B), and the white arrow indicates the posterior bulb. (C and D) Tail region, L1 larvae. Black arrow indicates the rectum (missing in D), and the white arrow indicates the intestine. Note the distension of the intestine in D resulting from the absence of the rectum. (E and F) Embryos 6–7 h after egg laying. (E) The embryo has reached the 3-fold stage. (F) The embryo has arrested without undergoing elongation. We also observed embryos with the same overall morphology as in F when we injected N2 hermaphrodites with a mixture of glp-1 and lin-12 dsRNAs, or with lag-1 dsRNA.
Figure 4
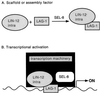
Models for SEL-8 function. SEL-8 is a nuclear protein that is required for LIN-12 and GLP-1 signaling. Although several models would be consistent with these basic observations, we feature two here. (A) Scaffold/assembly factor. SEL-8 may serve to stabilize or to facilitate the assembly of a complex that contains LIN-12(intra)/GLP-1(intra) and LAG-1, and perhaps other proteins. (B) Transcriptional activation. The glutamine-rich regions of SEL-8 may function as a transcriptional activation domain that interacts with proteins in the RNA polymerase II transcription initiation complex.
Similar articles
- p24 proteins and quality control of LIN-12 and GLP-1 trafficking in Caenorhabditis elegans.
Wen C, Greenwald I. Wen C, et al. J Cell Biol. 1999 Jun 14;145(6):1165-75. doi: 10.1083/jcb.145.6.1165. J Cell Biol. 1999. PMID: 10366590 Free PMC article. - lag-1, a gene required for lin-12 and glp-1 signaling in Caenorhabditis elegans, is homologous to human CBF1 and Drosophila Su(H).
Christensen S, Kodoyianni V, Bosenberg M, Friedman L, Kimble J. Christensen S, et al. Development. 1996 May;122(5):1373-83. doi: 10.1242/dev.122.5.1373. Development. 1996. PMID: 8625826 - sel-10, a negative regulator of lin-12 activity in Caenorhabditis elegans, encodes a member of the CDC4 family of proteins.
Hubbard EJ, Wu G, Kitajewski J, Greenwald I. Hubbard EJ, et al. Genes Dev. 1997 Dec 1;11(23):3182-93. doi: 10.1101/gad.11.23.3182. Genes Dev. 1997. PMID: 9389650 Free PMC article. - Notch signaling.
Artavanis-Tsakonas S, Matsuno K, Fortini ME. Artavanis-Tsakonas S, et al. Science. 1995 Apr 14;268(5208):225-32. doi: 10.1126/science.7716513. Science. 1995. PMID: 7716513 Review. - LIN-12/Notch signaling in C. elegans.
Greenwald I. Greenwald I. WormBook. 2005 Aug 8:1-16. doi: 10.1895/wormbook.1.10.1. WormBook. 2005. PMID: 18050403 Free PMC article. Review.
Cited by
- Redundant mechanisms regulating the proliferation vs. differentiation balance in the C. elegans germline.
Vanden Broek K, Han X, Hansen D. Vanden Broek K, et al. Front Cell Dev Biol. 2022 Sep 2;10:960999. doi: 10.3389/fcell.2022.960999. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 36120589 Free PMC article. Review. - Subunits of the DNA polymerase alpha-primase complex promote Notch-mediated proliferation with discrete and shared functions in C. elegans germline.
Yoon DS, Cha DS, Alfhili MA, Keiper BD, Lee MH. Yoon DS, et al. FEBS J. 2018 Jul;285(14):2590-2604. doi: 10.1111/febs.14512. Epub 2018 May 28. FEBS J. 2018. PMID: 29775245 Free PMC article. - Suppressors of the egg-laying defective phenotype of sel-12 presenilin mutants implicate the CoREST corepressor complex in LIN-12/Notch signaling in C. elegans.
Jarriault S, Greenwald I. Jarriault S, et al. Genes Dev. 2002 Oct 15;16(20):2713-28. doi: 10.1101/gad.1022402. Genes Dev. 2002. PMID: 12381669 Free PMC article. - Increasing Notch signaling antagonizes PRC2-mediated silencing to promote reprograming of germ cells into neurons.
Seelk S, Adrian-Kalchhauser I, Hargitai B, Hajduskova M, Gutnik S, Tursun B, Ciosk R. Seelk S, et al. Elife. 2016 Sep 7;5:e15477. doi: 10.7554/eLife.15477. Elife. 2016. PMID: 27602485 Free PMC article. - Nrarp is a novel intracellular component of the Notch signaling pathway.
Lamar E, Deblandre G, Wettstein D, Gawantka V, Pollet N, Niehrs C, Kintner C. Lamar E, et al. Genes Dev. 2001 Aug 1;15(15):1885-99. doi: 10.1101/gad.908101. Genes Dev. 2001. PMID: 11485984 Free PMC article.
References
- Greenwald I. Genes Dev. 1998;12:1751–1762. - PubMed
- Brou C, Logeat F, Gupta N, Bessia C, LeBail O, Doedens J R, Cumano A, Roux P, Black R A, Israel A. Mol Cell. 2000;5:207–216. - PubMed
- Mumm J S, Schroeter E H, Saxena M T, Griesemer A, Xu T, Pan D J, Ray W J, Kopan R. Mol Cell. 2000;5:197–206. - PubMed
- Schroeter E H, Kisslinger J A, Kopan R. Nature (London) 1998;393:382–386. - PubMed
- Struhl G, Adachi A. Cell. 1998;93:649–660. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases