Nucleus-vacuole junctions in Saccharomyces cerevisiae are formed through the direct interaction of Vac8p with Nvj1p - PubMed (original) (raw)
Nucleus-vacuole junctions in Saccharomyces cerevisiae are formed through the direct interaction of Vac8p with Nvj1p
X Pan et al. Mol Biol Cell. 2000 Jul.
Free PMC article
Abstract
Vac8p is a vacuolar membrane protein that is required for efficient vacuole inheritance and fusion, cytosol-to-vacuole targeting, and sporulation. By analogy to other armadillo domain proteins, including beta-catenin and importin alpha, we hypothesize that Vac8p docks various factors at the vacuole membrane. Two-hybrid and copurfication assays demonstrated that Vac8p does form complexes with multiple binding partners, including Apg13p, Vab2p, and Nvj1p. Here we describe the surprising role of Vac8p-Nvj1p complexes in the formation of nucleus-vacuole (NV) junctions. Nvj1p is an integral membrane protein of the nuclear envelope and interacts with Vac8p in the cytosol through its C-terminal 40-60 amino acids (aa). Nvj1p green fluorescent protein (GFP) concentrated in small patches or rafts at sites of close contact between the nucleus and one or more vacuoles. Previously, we showed that Vac8p-GFP concentrated in intervacuole rafts, where is it likely to facilitate vacuole-vacuole fusion, and in "orphan" rafts at the edges of vacuole clusters. Orphan rafts of Vac8p red-sifted GFP (YFP) colocalize at sites of NV junctions with Nvj1p blue-sifted GFP (CFP). GFP-tagged nuclear pore complexes (NPCs) were excluded from NV junctions. In vac8-Delta cells, Nvj1p-GFP generally failed to concentrate into rafts and, instead, encircled the nucleus. NV junctions were absent in both nvj1-Delta and vac8-Delta cells. Overexpression of Nvj1p caused the profound proliferation of NV junctions. We conclude that Vac8p and Nvj1p are necessary components of a novel interorganelle junction apparatus.
Figures
Figure 1
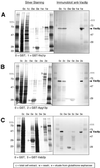
Glutathione–Sepharose affinity copurification of Vac8p with three binding partners. Proteins from cell extracts (c), wash solutions (w), and eluates (e) were separated by SDS-PAGE and were analyzed by silver staining (left panels) and immunoblot using anti-Vac8p antibodies (right panels). (A) Copurification of Vac8p with GST-Nvj1p. (B) Copurification of Vac8p with GST-Apg13p. (C) Copurification of Vac8p with GST-Vab2p. Labeled bands correspond to GST (27 kDa), GST-Nvj1p (63.4 kDa), and Vac8p (63.2 kDa).
Figure 2
Nvj1p is an integral membrane protein. (A) Hydrophobicity plot and map of Nvj1p. The map shows the location of the putative N-terminal signal peptide (M1TRPPLV_R_G_IFSLGLSVAVL_KGVEK25), the significant hydrophobic region (V96LILLSFLLPIAWTVL112), and the portions of NVJ1 that were isolated in independent two-hybrid clones (aa 93–321 and 119–321). (B) Nvj1p fractionates as an integral membrane protein. Panel 1: A whole cell extract from cells expressing Nvj1p-GFP was separated into supernatant (S100) and membrane (P100) fraction by centrifugation. P100 was subsequently treated as described in Material and Methods (S, supernatant; P, pellet). The presence of Nvj1p in each fraction was assayed by immunoblot using anti-GFP antibodies. Panel 2: Extract from cells expressing GFP-Nvj1p and Vac8p were treated as described in panel 1.
Figure 3
Two hybrid mapping of the C-terminal domain of Nvj1p that interacts with Vac8p.
Figure 4
API processing is normal in nvj1-Δ cells. Midlog-phase cells were incubated for 4 h in rich (YEPD) or nitrogen starvation (SD-N) medium. Cell lysates were prepared and processed for immunoblotting with anti-API antibody. The positions of precursor and mature API are indicated.
Figure 5
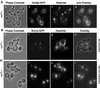
Nvj1p-GFP localizes to a specialized region of the NE in a _VAC8_-dependent manner. (A) Localization of cNvj1p-GFP in wt cells grown to an OD600 of 2.8 in SCGlu (panels a and b), and in cells grown to an OD600 of 10 in YPD and costained for DNA (Hoechst) (panel c) and vacuoles (FM4–64) (panel d). (B) Nvj1p-GFP in wt (VAC8+) (panels a and b) and vac8-Δ cells (panels d and e) grown in SC medium to OD600 ∼0.1–0.3. Localization of Nvj1p-GFP in wt and vac8-Δ cells stained for DNA (Hoechst) and vacuoles (FM4–64) (panels c and f). Plasmid-borne Nvj1p-GFP was expressed uninduced from the CUP1 promoter.
Figure 6
Localization of Vac8p-GFP and Nvj1p-GFP to single perinuclear rafts in vac7-Δ cells. Vac8p-GFP localization (A), Nvj1p-GFP localization before (B) and after (C) the induction of expression by 0.1 mM CuSO4. Phase contrast (panels a, e, and i), GFP (panels b, f, and j), Hoecht (panels c, g, and k), and overlay of phase contrast and Hoechst images (panels d, h, and l). Bar: 5 μm.
Figure 7
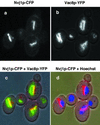
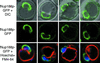
Partial colocalization of Vac8p and Nvj1p. Nvj1p-CFP (blue-shifted GFP) and Vac8p-YFP (red-shifted GFP) were localized in the same cells. Monochromic images of Nvj1p-CFP (a) and Vac8p-YFP (b), and color overlay images over a DIC mask of Nvj1p-CFP and Vac8p-YFP (c), and Nvj1p-CFP, Hoechst, and DIC (d). In panel c, yellow occurs where Nvj1p-CFP and Vac8p-YFP overlap.
Figure 8
NVJ1 expression levels control the occurrence and morphology of NV junctions. (A) wt cells + empty vector; (B) nvj1-Δ cells + empty vector; (C) wt cells + PCUP1_-NVJ1_; (D) wt cells + PCUP1_-_-_NVJ1_-GFP. Overexpression from the CUP1 promoter was achieved by adding 0.1 mM CuSO4 to growth media for 3 h. Cells were subsequently fixed and prepared for TEM as described in Material and Methods.
Figure 9
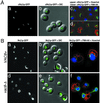
Nup188p-GFP-tagged NPCs are excluded from NV junctions. Pore complexes in nup188-Δ cells were labeled with Nup188p-GFP. Vacuoles were stained with FM4–64, and chromatin with Hoechst (Materials and Methods). Examples of four cells that exhibited large NV junctions are shown. Upper panels: Overlays of GFP-Nup188p and DIC images. Middle panels: GFP-Nup188p. Lower panels: Overlays of GFP-Nup188p, FM4–64, and Hoechst.
Similar articles
- Nvj1p is the outer-nuclear-membrane receptor for oxysterol-binding protein homolog Osh1p in Saccharomyces cerevisiae.
Kvam E, Goldfarb DS. Kvam E, et al. J Cell Sci. 2004 Oct 1;117(Pt 21):4959-68. doi: 10.1242/jcs.01372. Epub 2004 Sep 14. J Cell Sci. 2004. PMID: 15367582 - Mechanistic insight into the nucleus-vacuole junction based on the Vac8p-Nvj1p crystal structure.
Jeong H, Park J, Kim HI, Lee M, Ko YJ, Lee S, Jun Y, Lee C. Jeong H, et al. Proc Natl Acad Sci U S A. 2017 Jun 6;114(23):E4539-E4548. doi: 10.1073/pnas.1701030114. Epub 2017 May 22. Proc Natl Acad Sci U S A. 2017. PMID: 28533415 Free PMC article. - Targeting of Tsc13p to nucleus-vacuole junctions: a role for very-long-chain fatty acids in the biogenesis of microautophagic vesicles.
Kvam E, Gable K, Dunn TM, Goldfarb DS. Kvam E, et al. Mol Biol Cell. 2005 Sep;16(9):3987-98. doi: 10.1091/mbc.e05-04-0290. Epub 2005 Jun 15. Mol Biol Cell. 2005. PMID: 15958487 Free PMC article. - Nucleus-vacuole junctions and piecemeal microautophagy of the nucleus in S. cerevisiae.
Kvam E, Goldfarb DS. Kvam E, et al. Autophagy. 2007 Mar-Apr;3(2):85-92. doi: 10.4161/auto.3586. Epub 2007 Mar 2. Autophagy. 2007. PMID: 17204844 Review. - Nucleus-vacuole junctions in yeast: anatomy of a membrane contact site.
Kvam E, Goldfarb DS. Kvam E, et al. Biochem Soc Trans. 2006 Jun;34(Pt 3):340-2. doi: 10.1042/BST0340340. Biochem Soc Trans. 2006. PMID: 16709156 Review.
Cited by
- Autophagic processes in yeast: mechanism, machinery and regulation.
Reggiori F, Klionsky DJ. Reggiori F, et al. Genetics. 2013 Jun;194(2):341-61. doi: 10.1534/genetics.112.149013. Genetics. 2013. PMID: 23733851 Free PMC article. Review. - Lifelong dietary protein restriction accelerates skeletal muscle loss and reduces muscle fibre size by impairing proteostasis and mitochondrial homeostasis.
Ersoy U, Kanakis I, Alameddine M, Pedraza-Vazquez G, Ozanne SE, Peffers MJ, Jackson MJ, Goljanek-Whysall K, Vasilaki A. Ersoy U, et al. Redox Biol. 2024 Feb;69:102980. doi: 10.1016/j.redox.2023.102980. Epub 2023 Dec 2. Redox Biol. 2024. PMID: 38064763 Free PMC article. - Yeast Vps13 promotes mitochondrial function and is localized at membrane contact sites.
Park JS, Thorsness MK, Policastro R, McGoldrick LL, Hollingsworth NM, Thorsness PE, Neiman AM. Park JS, et al. Mol Biol Cell. 2016 Aug 1;27(15):2435-49. doi: 10.1091/mbc.E16-02-0112. Epub 2016 Jun 8. Mol Biol Cell. 2016. PMID: 27280386 Free PMC article. - Rapid cytoplasmic turnover of yeast ribosomes in response to rapamycin inhibition of TOR.
Pestov DG, Shcherbik N. Pestov DG, et al. Mol Cell Biol. 2012 Jun;32(11):2135-44. doi: 10.1128/MCB.06763-11. Epub 2012 Mar 26. Mol Cell Biol. 2012. PMID: 22451491 Free PMC article. - Endoplasmic Reticulum-Vacuole Contact Sites "Bloom" With Stress-Induced Lipid Droplets.
Henne WM, Hariri H. Henne WM, et al. Contact (Thousand Oaks). 2018 Jan-Dec;1:10.1177/2515256418756112. doi: 10.1177/2515256418756112. Epub 2018 Apr 3. Contact (Thousand Oaks). 2018. PMID: 30112463 Free PMC article.
References
- Achleitner G, Gaigg B, Krasser A, Kainersdorfer E, Kohlwein SD, Perktold A, Zellnig G, Daum G. Association between endoplasmic reticulum and mitochondria of yeast facilitates interorganelle transport of phospholipids through membrane contact. Eur J Biochem. 1999;264:545–553. - PubMed
- Bartel PL, Fields S. Analyzing protein-protein interactions using two-hybrid system. Methods Enzymol. 1995;254:241–263. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous