Precursors to the U3 small nucleolar RNA lack small nucleolar RNP proteins but are stabilized by La binding - PubMed (original) (raw)
Precursors to the U3 small nucleolar RNA lack small nucleolar RNP proteins but are stabilized by La binding
J Kufel et al. Mol Cell Biol. 2000 Aug.
Abstract
Almost all small eukaryotic RNAs are processed from transiently stabilized 3'-extended forms. A key question is how and why such intermediates are stabilized and how they can then be processed to the mature RNA. Here we report that yeast U3 is also processed from a 3'-extended precursor. The major 3'-extended forms of U3 (U3-3'I and -II) lack the cap trimethylation present in mature U3 and are not associated with small nucleolar RNP (snoRNP) proteins that bind mature U3, i.e., Nop1p, Nop56p, and Nop58p. Depletion of Nop58p leads to the loss of mature U3 but increases the level of U3-3'I and -II, indicating a requirement for the snoRNP proteins for final maturation. Pre-U3 is cleaved by the endonuclease Rnt1p, but U3-3'I and -II do not extend to the Rnt1p cleavage sites. Rather, they terminate at poly(U) tracts, suggesting that they might be bound by Lhp1p (the yeast homologue of La). Immunoprecipitation of Lhp1p fused to Staphylococcus aureus protein A resulted in coprecipitation of both U3-3'I and -II. Deletion of LHP1, which is nonessential, led to the loss of U3-3'I and -II. We conclude that pre-U3 is cleaved by Rnt1p, followed by exonuclease digestion to U3-3'I and -II. These species are stabilized against continued degradation by binding of Lhp1p. Displacement of Lhp1p by binding of the snoRNP proteins allows final maturation, which involves the exosome complex of 3'-->5' exonucleases.
Figures
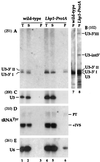
FIG. 1
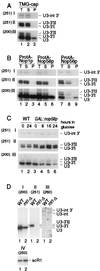
Northern analysis of 3′-extended forms of U3 snoRNA. Probes (indicated in parentheses): 251, complementary to the region across the 3′ end of the mature U3A; 200, complementary to mature U3; 260, complementary to the U3A intron; 250, complementary to the scR1 RNA. For panels A and B, input lysates were estimated to contain comparable amounts of U3 snoRNA, and equal fractions of the preparation were loaded for each lane; panels C and D, constant amounts of total RNA were loaded in each lane. (A) Immunoprecipitation with m32,2,7G cap-specific antibody (R1131) on lysates from the wild-type D150 strain. (B) Immunoprecipitation of lysates from strains expressing epitope-tagged fusion proteins ProtA-Nop1p, ProtA-Nop58p, and ProtA-Nop56p. (C) Stability of mature and 3′-extended U3 upon depletion of Nop58p. RNA was extracted from the GAL::nop58 and wild-type (WT) strains following transfer from permissive, galactose medium to repressive, glucose medium for the times indicated. (D) Effects of _rnt1_-Δ on 3′-extended U3. The level of scR1 RNA is shown as a control for loading. T, total cell lysate; S, immune supernatant; P, immunoprecipitate.
FIG. 2
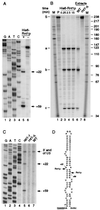
Rnt1p cleaves the 3′ end of the U3 precursor. (A) Mapping of the in vitro Rnt1p cleavage sites. Primer extension was performed with probe 253 on the model U3(−60/+139) RNA incubated with buffer (lane 6) or recombinant His6-Rnt1p (lane 5) as described in Materials and Methods. DNA sequencing reaction on a PCR product encompassing the 3′ end of U3 from positions −60 to +139, using the same primer, was run in parallel (lanes 1 to 4). The primer extension stops at positions +22 and +59 are indicated. (B) In vitro cleavage of an internally labeled model U3(−60/+177) RNA substrate by Rnt1p. 32P-labeled U3(−60/+177) RNA was incubated at 23°C in the following conditions: lane 2, Rnt1p buffer; lanes 3 to 5, Rnt1p buffer with 10 ng of recombinant His6-Rnt1p for the times indicated; lane 6, with whole-cell extract from a wild-type (WT) strain of yeast; lane 7, with whole-cell extract from an _rnt1_-Δ strain. Lanes 1 and 8, RNA size markers. The positions of DNA size markers are indicated on the right in nucleotides. The obtained cleavage products are labeled a to c on the left, and the predicted origins of these species are as follows: S, substrate (237 nt); a, 3′ end of transcript to position +21/+22 (119 nt); b, 5′ end of transcript to position +58/+59 (81 nt); c, positions +21/+22 to +58/+59 (37 nt). Since in vitro cleavages of U3(−60/+177) are complete (100%), no intermediate cleav- age products are visible. (C) Mapping of the Rnt1p 5′ cleavage site in vitro. Primer extension analysis through the 3′ end of the pre-U3 was performed with primer 252, hybridizing downstream of position +177. RNA was extracted from wild-type (lane 7) and _rnt1_-Δ (lane 6) strains grown at 30°C and from a rat1-1 strain following transfer to 37°C for 2 h (lane 5). DNA sequencing reactions were run in parallel (lanes 1 to 4). The primer extension stops at positions +59, +22, and +1 (3′ end of U3) are indicated. (D) Computer-predicted RNA structure in the U3 3′ flanking region that contains the Rnt1p cleavage sites. The cleavage sites between nt +21 and +22 and between nt +58 and +59 are indicated by arrows. The 3′ end of mature U3 is underlined.
FIG. 3
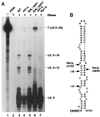
Mapping of the 3′-extended forms of U3 by RNase protection. (A) RNA was extracted from wild-type (WT), _rnt1_-Δ, and _lhp1_-Δ strains grown at 30°C and from GAL::rrp41 and _GAL::rrp41/rnt1_-Δ strains following transfer from permissive, RSG medium to repressive, glucose medium at 30°C for 24 and 48 h, respectively. Total E. coli tRNA was used as a control RNA. Positions of the Rnt1p-dependent protected species at +12 and +18 are indicated. (B) Schematic of the U3 3′ flanking region showing the ends of the protected regions and the Rnt1p cleavage sites.
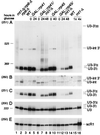
FIG. 4
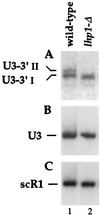
3′-extended forms of U3 are stabilized by Lhp1p. Lane 1, LHP1 strain; lane 2, _lhp1_-Δ strain. Total RNA was analyzed by Northern hybridization with probe 251, specific for the 3′-extended U3 (A), probe 200, which hybridizes to the mature U3 (B), and probe 250, which hybridizes to scR1 RNA (C).
FIG. 5
3′-extended forms of U3 are coprecipitated with Lhp1p-ProtA. Lysates from the LHP1+ and LHP1::ProtA strains were immunoprecipitated using IgG agarose. RNA was recovered from the total cell lysate (T), immune supernatant (S), and immunoprecipitate (P) and analyzed by Northern hybridization. Probes are indicated in parentheses and described in Materials and Methods. On prolonged exposure, background precipitation of mature U3 is seen for both the wild-type and Lhp1-ProtA strains (lanes 7 and 8). In panel B, the total and supernatant lanes were heavily overexposed at the exposure needed to visualize the U3-int 3′ and U3-3′III RNAs and were omitted. Approximately fourfold more cell equivalents are loaded for the bound material.
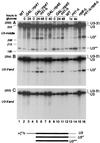
FIG. 6
Northern analysis of processing of U3 snoRNA in exosome mutants. RNA was extracted from strains carrying _GAL_-regulated constructs following transfer from permissive, RSG medium to repressive, glucose medium at 30°C for the times indicated, or from the wild-type (WT), _rnt1_-Δ, _rrp6_-Δ, and rnt1-Δ/rrp6-Δ strains grown on glucose medium at 30°C. RNA was separated on an 6% polyacrylamide gel and hybridized with oligonucleotide probes. The panels show successive hybridization of the same filter. Probes are indicated in parentheses on the left and described in Materials and Methods; the positions of RNA species detected are indicated on the right. Panel C presents a weaker exposure of the same gel as panel A. Panels B to E present only relevant regions of the Northern blots. The amount of total RNA loaded in lane 16 is fourfold greater than in lane 15 and other lanes. The positions of migration of scR1 (525 nt) and P (369 nt) RNAs determined by hybridization of the same filter are indicated as size markers. Mature U3 is 333 nt.
FIG. 7
Exosome components participate in the degradation of U3 snoRNA. For Northern analysis of U3 snoRNA in wild-type (WT) and _rnt1_-Δ and exosome mutant strains. RNA was extracted as described for Fig. 2, separated on an 6% polyacrylamide gel, and hybridized with oligonucleotide probes. The panels show successive hybridization of the same filter. Probes are indicated in parentheses on the left and described in Materials and Methods; the positions of RNA species detected are indicated on the right. The amount of total RNA loaded in lane 14 is fourfold greater than in lane 13 and other lanes. The positions of migration of snRNA190 (190 nt), U5L (215 nt), and snR10 (246 nt) determined by hybridization of the same filter are indicated as size markers. Mature U3 is 333 nt. The locations of the oligonucleotide probes and the predicted structures of the degradation intermediates are shown schematically.
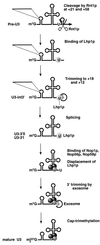
FIG. 8
Model for the 3′ processing of the U3A snoRNA. The presence of the poly(U) tracts and stem-loop structure in the 3′ flanking sequence and the intron are indicated. For simplicity, only one poly(U) tract is indicated. In reality, two tracts are present, at +19 and +13, each of which is likely to act as a binding site for Lhp1p. The activity that carries out the initial trimming to +18 and +12 has not been determined but is likely to be the exosome. The endpoints of the U3-int 3′ species have not been determined, but the finding that these species are associated with Lhp1p suggests that they are processed to +18 and +12.
FIG. 9
Comparison of the 3′ flanking sequence of U3A to those of the U1, U2, U4, and U5 snRNAs. In panel A, the Rnt1p cleavage sites (\) have been aligned. The mature regions of U3, U2, and U5L are in uppercase. For U1, U4, and U5S, the mature regions are further from the Rnt1p cleavage site. These are aligned in the panel B. Poly(U) sequences of four or more residues are underlined.
Similar articles
- Implication of the box C/D snoRNP assembly factor Rsa1p in U3 snoRNP assembly.
Rothé B, Manival X, Rolland N, Charron C, Senty-Ségault V, Branlant C, Charpentier B. Rothé B, et al. Nucleic Acids Res. 2017 Jul 7;45(12):7455-7473. doi: 10.1093/nar/gkx424. Nucleic Acids Res. 2017. PMID: 28505348 Free PMC article. - A complex pathway for 3' processing of the yeast U3 snoRNA.
Kufel J, Allmang C, Verdone L, Beggs J, Tollervey D. Kufel J, et al. Nucleic Acids Res. 2003 Dec 1;31(23):6788-97. doi: 10.1093/nar/gkg904. Nucleic Acids Res. 2003. PMID: 14627812 Free PMC article. - Yeast exosome mutants accumulate 3'-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs.
van Hoof A, Lennertz P, Parker R. van Hoof A, et al. Mol Cell Biol. 2000 Jan;20(2):441-52. doi: 10.1128/MCB.20.2.441-452.2000. Mol Cell Biol. 2000. PMID: 10611222 Free PMC article. - Sno-capped: 5' ends of preribosomal RNAs are decorated with a U3 SnoRNP.
Culver GM. Culver GM. Chem Biol. 2002 Jul;9(7):777-9. doi: 10.1016/s1074-5521(02)00171-0. Chem Biol. 2002. PMID: 12144919 Review. - SnoRNP biogenesis meets Pre-mRNA splicing.
Kiss T. Kiss T. Mol Cell. 2006 Sep 15;23(6):775-6. doi: 10.1016/j.molcel.2006.08.023. Mol Cell. 2006. PMID: 16973429 Review.
Cited by
- RNA chaperone activity of human La protein is mediated by variant RNA recognition motif.
Naeeni AR, Conte MR, Bayfield MA. Naeeni AR, et al. J Biol Chem. 2012 Feb 17;287(8):5472-82. doi: 10.1074/jbc.M111.276071. Epub 2011 Dec 27. J Biol Chem. 2012. PMID: 22203678 Free PMC article. - CK2 is responsible for phosphorylation of human La protein serine-366 and can modulate rpL37 5'-terminal oligopyrimidine mRNA metabolism.
Schwartz EI, Intine RV, Maraia RJ. Schwartz EI, et al. Mol Cell Biol. 2004 Nov;24(21):9580-91. doi: 10.1128/MCB.24.21.9580-9591.2004. Mol Cell Biol. 2004. PMID: 15485924 Free PMC article. - The mammalian exosome mediates the efficient degradation of mRNAs that contain AU-rich elements.
Mukherjee D, Gao M, O'Connor JP, Raijmakers R, Pruijn G, Lutz CS, Wilusz J. Mukherjee D, et al. EMBO J. 2002 Jan 15;21(1-2):165-74. doi: 10.1093/emboj/21.1.165. EMBO J. 2002. PMID: 11782436 Free PMC article. - Small nucleolar RNAs: versatile trans-acting molecules of ancient evolutionary origin.
Terns MP, Terns RM. Terns MP, et al. Gene Expr. 2002;10(1-2):17-39. Gene Expr. 2002. PMID: 11868985 Free PMC article. Review. - The Hsp90 chaperone controls the biogenesis of L7Ae RNPs through conserved machinery.
Boulon S, Marmier-Gourrier N, Pradet-Balade B, Wurth L, Verheggen C, Jády BE, Rothé B, Pescia C, Robert MC, Kiss T, Bardoni B, Krol A, Branlant C, Allmang C, Bertrand E, Charpentier B. Boulon S, et al. J Cell Biol. 2008 Feb 11;180(3):579-95. doi: 10.1083/jcb.200708110. J Cell Biol. 2008. PMID: 18268104 Free PMC article.
References
- Abou Elela S, Igel H, Ares M., Jr RNase III cleaves eukaryotic preribosomal RNA at a U3 snoRNP-dependent site. Cell. 1996;85:115–124. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous