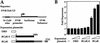BCoR, a novel corepressor involved in BCL-6 repression - PubMed (original) (raw)
. 2000 Jul 15;14(14):1810-23.
Affiliations
- PMID: 10898795
- PMCID: PMC316791
BCoR, a novel corepressor involved in BCL-6 repression
K D Huynh et al. Genes Dev. 2000.
Abstract
BCL-6 encodes a POZ/zinc finger transcriptional repressor that is required for germinal center formation and may influence apoptosis. Aberrant expression of BCL-6 due to chromosomal translocations is implicated in certain subtypes of non-Hodgkin's lymphoma. The POZ domains of BCL-6 and several other POZ proteins interact with corepressors N-CoR and SMRT. Here we identify and characterize a novel corepressor BCoR (BCL-6 interacting corepressor), which is expressed ubiquitously in human tissues. BCoR can function as a corepressor when tethered to DNA and, when overexpressed, can potentiate BCL-6 repression. Specific class I and II histone deacetylases (HDACs) interact in vivo with BCoR, suggesting that BCoR may functionally link these two classes of HDACs. Strikingly, BCoR interacts selectively with the POZ domain of BCL-6 but not with eight other POZ proteins tested, including PLZF. Additionally, interactions between the BCL-6 POZ domain and SMRT, N-CoR, and BCoR are mutually exclusive. The specificity of the BCL-6/BCoR interaction suggests that BCoR may have a role in BCL-6-associated lymphomas.
Figures
Figure 1
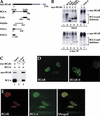
Amino acid sequence and tissue distribution of BCoR. (A) Deduced amino acid sequence of BCoR. The point of divergence between BCoR and BCoR-S is indicated (▾); the three tandem ankyrin repeats are underlined. (B) Schematic diagram of BCoR and BCoR-S. The ankyrin repeats (codons 1428–1527) and the region corresponding to the yeast two-hybrid-isolated cDNA clone (A3; codons 112–753) are indicated. (▾) The point of divergence between the two proteins. (C) Expression of BCoR mRNA in various human tissues. A Human RNA Master Blot (Clontech) was probed with a cDNA fragment derived from A3. Tissue identification grid, courtesy of Clontech.
Figure 2
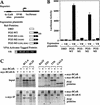
Full-length BCL-6 and BCoR interact in vitro and in vivo, the POZ domain of BCL-6 is both necessary and sufficient for interaction with BCoR in vitro, and BCL-6 and BCoR colocalize in the nucleus. (A) Structure of the proteins used in B and C. (Solid oval) The POZ domain; (solid rectangle) zinc fingers. (*) The myc epitope tag. (B) Coimmunoprecipitation of BCL-6 and BCoR in vitro. Pair-wise combinations of [35S]methionine-labeled nontagged BCL-6 derivatives and myc-tagged BCoR were produced by cotranslation and immunoprecipitated with the α-myc antibody. The immune complexes were recovered and analyzed by SDS-PAGE (10% for lanes 1–5; 17% for lanes 6,7). The intensity of myc–BCoR in lanes 6 and 7 appears stronger due to compression on the 17% gel of non-full-length myc-BCoR proteins. Input represents 20% of the total used. (●) The expected migration of the nontagged BCL-6 derivatives. (C) Coimmunoprecipitation of BCL-6 and BCoR in vivo. 293 cells were transfected with 3 μg of BCL-6 expression plasmid alone (lane 2) or cotransfected with 3 μg each of BCL-6 and myc-tagged BCoR expression plasmids (lanes 1,3). Cell lysates were immunoprecipitated with α-myc antibody (lanes 2,3) and the recovered proteins were detected by Western blot analysis using N-3 α-BCL-6 and α-myc antibodies. The input (lane 1) corresponds to 1/50 of the lysate used in the immunoprecipitation (lane 3). (D) Subcellular localization of BCoR and BCoR-S. Immunofluorescence was performed on HeLa cells transfected with 1.2 μg of either myc-tagged BCoR or BCoR-S expression plasmids. Multiple Z series confocal microscopy images of the given fields were compiled for each image of BCoR and BCoR-S. (E) Colocalization of BCL-6 and BCoR. Coimmunofluorescence was performed on HeLa cells cotransfected with 0.6 μg each of BCL-6 and myc-tagged BCoR, and the data collected as in D. The compiled images of BCoR and BCL-6 were merged to detect colocalization.
Figure 3
Self association of the POZ domain of BCL-6 is required for interaction with BCoR, and BCoR interacts selectively with the POZ domain of BCL-6. (A) Structures of the reporter construct, bait, and activator proteins used in B. The bait proteins consist of the GAL4 DBD alone, or fused to wild type or the indicated mutant POZ domains. The activator-tagged protein VB consists of the VP16 activation domain fused to the yeast two-hybrid-isolated BCoR clone (codons 112–753). (B) Interactions of BCoR (codons 112–753) with BCL-6 POZ domain point mutants in the mammalian two-hybrid assay. Luciferase and β-gal assays were conducted on cell extracts from HeLa cells transiently transfected with 500 ng of the GAL4-responsive reporter (pAH205), 50 ng of the indicated bait expression plasmids; 0, 50, or 250 ng of the VB expression plasmid; and 25 ng of the pCMVlacZ plasmid, as control for transfection efficiency. Data displayed represent the average of two experiments with the standard deviations depicted. (C) In vitro coimmunoprecipitation of BCoR and BCoR-S with POZ-containing proteins. The indicated pair-wise combinations of [35S]methionine-labeled nontagged POZ proteins (BCL-6, PLZF, BAZF, ZID, TTK, and GAGA) and either myc-tagged BCoR or BCoR-S were produced by cotranslation and immunoprecipitated with α-myc antibody. The proteins were recovered and analyzed by 8% SDS-PAGE. (●) The expected migration of the nontagged BCL-6 derivatives.
Figure 4
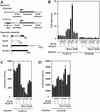
BCoR is a transcriptional corepressor. (A) Structures of the reporter construct and proteins used in B. (B) Luciferase and β-gal assays were performed on cell extracts from HeLa cells transiently transfected with 50 ng of the GAL4-responsive reporter (SV40 Enh G5), along with 0, 5, 10, or 25 ng of the indicated GAL4 fusion constructs [GAL4 DBD alone (DBD) or DBD fused with BCoR or BCoR-S), and 25 ng of pCMVLacZ plasmid. Data displayed represent the average of two experiments, with the standard deviations depicted. Fold repression was determined by normalizing luciferase/β-gal values of all samples to that of the reporter alone.
Figure 5
BCoR can selectively potentiate BCL-6 repression. (A) Structures of the reporter constructs and proteins used in B. (B) Luciferase and β-gal assays were conducted on HeLa cell extracts transiently transfected with 250 ng of either the SV40 Enh or 5× BCL-6 reporter construct, and the indicated combinations of BCL-6 (25 ng), BAZF (25 ng), BCoR (25 and 50 ng), and BCoR-S (25 and 50 ng); pEFLacZ (200 ng) was also included as control for transfection efficiency. Data displayed represent the average of two experiments with the standard deviations depicted. Fold repression was determined by normalizing luciferase/β-gal values (C,D) of all samples to that of the SV40 Enh reporter alone.
Figure 6
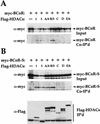
BCoR and BCoR-S interact with class I and class II HDACs in vivo. 293 cells were transfected with 4 μg of either myc-tagged BCoR (A) or BCoR-S (B) expression plasmids alone (first lane), or cotransfected with pair-wise combinations of 3 μg of the indicated Flag-tagged HDACs and 3 μg of either myc-tagged BCoR (A) or BCoR-S (B) expression plasmids (remaining lanes). Cell lysates were imunoprecipitated with α-Flag antibody, and the recovered proteins were detected by Western blot analysis using α-myc and α-Flag antibodies (shown in B only). The input corresponds to 1/100 of the lysates used for coimmunoprecipitation.
Figure 7
Interactions between the BCL-6 POZ domain and N-CoR, SMRT, and BCoR are mutually exclusive. (A) Structures of the reporter construct and proteins used in B. (B) Luciferase and β-gal assays were performed on HeLa cell extracts transiently transfected with 250 ng of the GAL4-responsive reporter (pAH205), 2.5 ng of the POZ bait, and the indicated combinations of VP16-tagged (VS, 25 ng; VN, 25 ng; VA, 5 ng) and nontagged (S, N, A, at 100 ng each) corepressor expression plasmids, along with 25 ng of the pCMVlacZ plasmid. Data displayed represent the average of two experiments with the standard deviations depicted.
Similar articles
- The BCL-6 POZ domain and other POZ domains interact with the co-repressors N-CoR and SMRT.
Huynh KD, Bardwell VJ. Huynh KD, et al. Oncogene. 1998 Nov 12;17(19):2473-84. doi: 10.1038/sj.onc.1202197. Oncogene. 1998. PMID: 9824158 - Critical residues within the BTB domain of PLZF and Bcl-6 modulate interaction with corepressors.
Melnick A, Carlile G, Ahmad KF, Kiang CL, Corcoran C, Bardwell V, Prive GG, Licht JD. Melnick A, et al. Mol Cell Biol. 2002 Mar;22(6):1804-18. doi: 10.1128/MCB.22.6.1804-1818.2002. Mol Cell Biol. 2002. PMID: 11865059 Free PMC article. - Class II histone deacetylases are directly recruited by BCL6 transcriptional repressor.
Lemercier C, Brocard MP, Puvion-Dutilleul F, Kao HY, Albagli O, Khochbin S. Lemercier C, et al. J Biol Chem. 2002 Jun 14;277(24):22045-52. doi: 10.1074/jbc.M201736200. Epub 2002 Apr 19. J Biol Chem. 2002. PMID: 11929873 - The LAZ3(BCL-6) oncoprotein recruits a SMRT/mSIN3A/histone deacetylase containing complex to mediate transcriptional repression.
Dhordain P, Lin RJ, Quief S, Lantoine D, Kerckaert JP, Evans RM, Albagli O. Dhordain P, et al. Nucleic Acids Res. 1998 Oct 15;26(20):4645-51. doi: 10.1093/nar/26.20.4645. Nucleic Acids Res. 1998. PMID: 9753732 Free PMC article. - Molecular pathogenesis of non-Hodgkin's lymphoma: the role of Bcl-6.
Pasqualucci L, Bereshchenko O, Niu H, Klein U, Basso K, Guglielmino R, Cattoretti G, Dalla-Favera R. Pasqualucci L, et al. Leuk Lymphoma. 2003;44 Suppl 3:S5-12. doi: 10.1080/10428190310001621588. Leuk Lymphoma. 2003. PMID: 15202519 Review.
Cited by
- The Relationship between RUVBL1 (Pontin, TIP49, NMP238) and BCL6 in Benign and Malignant Human Lymphoid Tissues.
Baron BW, Baron RM, Baron JM. Baron BW, et al. Biochem Biophys Rep. 2016 Jul;6:1-8. doi: 10.1016/j.bbrep.2016.02.006. Biochem Biophys Rep. 2016. PMID: 27066592 Free PMC article. - Bcl6a function is required during optic cup formation to prevent p53-dependent apoptosis and colobomata.
Lee J, Lee BK, Gross JM. Lee J, et al. Hum Mol Genet. 2013 Sep 1;22(17):3568-82. doi: 10.1093/hmg/ddt211. Epub 2013 May 12. Hum Mol Genet. 2013. PMID: 23669349 Free PMC article. - Novel Genetic Variations in Acute Myeloid Leukemia in Pakistani Population.
Shahid S, Shakeel M, Siddiqui S, Ahmed S, Sohail M, Khan IA, Abid A, Shamsi T. Shahid S, et al. Front Genet. 2020 Jun 23;11:560. doi: 10.3389/fgene.2020.00560. eCollection 2020. Front Genet. 2020. PMID: 32655615 Free PMC article. - Unbiased phosphoproteomic method identifies the initial effects of a methacrylic acid copolymer on macrophages.
Chamberlain MD, Wells LA, Lisovsky A, Guo H, Isserlin R, Talior-Volodarsky I, Mahou R, Emili A, Sefton MV. Chamberlain MD, et al. Proc Natl Acad Sci U S A. 2015 Aug 25;112(34):10673-8. doi: 10.1073/pnas.1508826112. Epub 2015 Aug 10. Proc Natl Acad Sci U S A. 2015. PMID: 26261332 Free PMC article. - Dynamic Changes in Gene Mutational Landscape With Preservation of Core Mutations in Mantle Cell Lymphoma Cells.
Zhang Q, Wang HY, Liu X, Roth MH, Shestov AA, Lee SC, Jain K, Soderquist C, Xiong QB, Ruella M, Strauser H, Glickson JD, Schuster SJ, Ptasznik A, Wasik MA. Zhang Q, et al. Front Oncol. 2019 Jul 3;9:568. doi: 10.3389/fonc.2019.00568. eCollection 2019. Front Oncol. 2019. PMID: 31334109 Free PMC article.
References
- Albagli O, Dhordain P, Deweindt C, Lecocq G, Leprince D. The BTB/POZ domain: A new protein-protein interaction motif common to DNA- and actin-binding proteins. Cell Growth Differ. 1995;6:1193–1198. - PubMed
- Albagli O, Dhordain P, Bernardin F, Quief S, Kerkaert JP, Leprince D. Multiple domains participate in distance-independent LAZ3/BCL6-mediated transcriptional repression. Biochem Biophys Res Commun. 1996;220:911–915. - PubMed
- Albagli O, Lantoine D, Quief S, Quignon F, Englert C, Kerckaert JP, Montarras D, Pinset C, Lindon C. Overexpressed BCL6 (LAZ3) oncoprotein triggers apoptosis, delays S phase progression and associates with replication foci. Oncogene. 1999;18:5063–5075. - PubMed
- Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials