The Ess1 prolyl isomerase is linked to chromatin remodeling complexes and the general transcription machinery - PubMed (original) (raw)
The Ess1 prolyl isomerase is linked to chromatin remodeling complexes and the general transcription machinery
X Wu et al. EMBO J. 2000.
Abstract
The Ess1/Pin1 peptidyl-prolyl isomerase (PPIase) is thought to control mitosis by binding to cell cycle regulatory proteins and altering their activity. Here we isolate temperature-sensitive ess1 mutants and identify six multicopy suppressors that rescue their mitotic-lethal phenotype. None are cell cycle regulators. Instead, five encode proteins involved in transcription that bind DNA, modify chromatin structure or are regulatory subunits of RNA polymerase II. A sixth suppressor, cyclophilin A, is a member of a distinct family of PPIases that are targets of immuno suppressive drugs. We show that the expression of some but not all genes is decreased in ess1 mutants, and that Ess1 interacts with the C-terminal domain (CTD) of RNA polymerase II in vitro and in vivo. The results forge a strong link between PPIases and the transcription machinery and suggest a new model for how Ess1/Pin1 controls mitosis. In this model, Ess1 binds and isomerizes the CTD of RNA polymerase II, thus altering its interaction with proteins required for transcription of essential cell cycle genes.
Figures
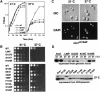
Fig. 1. Growth properties of _ess1_ts mutants. (A) Growth of wild-type (W303-1A) and selected mutants in rich medium (YEPD) at permissive and non-permissive temperatures. The _ess1_W15R allele was on a centromeric plasmid in an ess1Δ background. The _ess1_L94P and _ess1_H164R alleles were integrated into the genome. (B) Upper panel: growth of strains containing integrated _ess1_ts alleles. Cells were grown to mid-log phase in YEPD medium, and serial 1:5 dilutions were spotted onto plates and incubated for 1–2 days at the indicated temperatures. Lower panel: growth of strains carrying plasmid-borne (CEN) copies of _ess1_ts alleles in an ess1Δ host. Cells were grown and spotted as in the upper panel. (C) Mitotic arrest and nuclear fragmentation of _ess1_ts mutants. Cells (_ess1_H164R) were grown to mid-log phase and shifted to 37°C for 8 h. Cells were fixed, stained with DAPI, and visualized under Nomarski optics (DIC) or UV light (DAPI). (D) Western analysis of mutant proteins. Cells were grown at permissive temperature and shifted to 37°C for 4 h prior to harvesting. Equal amounts of total cell lysates were analyzed. Rabbit anti-Ess1 antiserum was used at a 1:4000 dilution.
Fig. 2. Identity of ts and lethal substitutions in Ess1. (A) Alignment of yeast Ess1 and homologs from Drosophila (Dodo) and human (Pin1), showing the position (underlined) and identity of the substitutions. (B) Model of the Ess1 structure based on X-ray crystallographic data of Pin1 (Ranganathan et al., 1997) showing the location of selected mutations. (C) Model of the active site of wild-type Ess1, C120R and H164R mutants, each containing a bound phosphoserine–proline dipeptide in the cis configuration. Although there are minor rearrangements, none appears to preclude binding of the substrate peptide.
Fig. 3. High-copy suppressors of the lethal ess1 mutant phenotype. (A) Suppression of _ess1_H164R host cells at 37°C by six different high-copy plasmids carrying the indicated genes. Control plasmids were YEpESS1 or an empty vector (pRS426). CaRPB7 is from Candida albicans. Cells were streaked onto medium that lacked uracil and incubated at 37°C for 5 days. (B) Fcp1 phosphatase activity is required for suppression of _ess1_ts mutants. Suppression of _ess1_H164R mutant cells at 37°C was tested using high-copy plasmids (2µ; TRP1) encoding wild-type (FCP1) or mutants alleles (fcp1-1 and fcp1-2) that lack CTD phosphatase activity (Kobor et al., 1999). (C) Western analysis to detect cyclophilin A (Cpr1) in whole-cell lysates from _ess1_H164R mutant cells carrying the indicated suppressors, or control plasmids, YEpESS1 or pRS426. Cells were grown at permissive temperature and shifted to 37°C for 4 h. Rabbit anti-cyclophilin A serum was used at a 1:3000 dilution.
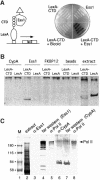
Fig. 4. Ess1 interacts with the CTD of RNA pol II in vivo and in vitro. (A) A schematic of the two-hybrid experiment is shown on the left. Yeast cells expressing the indicated proteins were streaked on an X-gal-containing plate and incubated overnight at 30°C (right). The LexA1–87–CTD bait protein contains nine repeats of the YSPTSPS motif and interacts with activation domain-tagged Ess1 but not with the control protein, Bicoid. Neither LexA1–87–CTD nor Ess1 alone activates transcription. (B) Ess1, but not cyclophilin A, interacts with LexA1–87–CTD in vitro. Cyclophilin A- (CypA), Ess1- or FKBP12-conjugated affinity beads or control beads were reacted with total protein extracts from cells expressing either a LexA1–87–CTD fusion protein or intact LexA1–202. Bound proteins were eluted and analyzed by western blotting with LexA antisera. Extract denotes whole-cell lysates not reacted with affinity beads. (C) Far-western analysis shows that Ess1 reacts with a protein that co-migrates with hyperphosphorylated RNA pol II. Lanes 1 and 2 are marker proteins and Coomassie-stained total yeast extracts, respectively. Lanes 4 and 7 are far-western samples using His-tagged Ess1 or CypA proteins as probes, and detected using the cognate antibodies. Lanes 3, 5, 6 and 8 are standard western blots reacted with the indicated antisera.
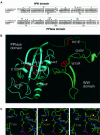
Fig. 5. ESS1 shows a genetic interaction with RPB1. (A) The _ess1_H164R mutant is synthetically lethal with a reduced dosage of the RPB1 gene. Cells of the indicated genotype (W303-1A background) were streaked to rich medium and grown for 3 days at permissive temperature (30°C). (B) The _ess1_H164R mutant is synthetically lethal with reduced levels of RPB1 expression. Cells of the indicated genotype (W303-1A background) were streaked to selective medium and grown for 3 days at permissive temperature (25°C). pRPB1 is a CEN-ARS (URA3) plasmid that expresses the large subunit of RNA pol II from its natural promoter. PLEU-RPB1 is a CEN-ARS (TRP1) plasmid (also known as pLEU-RPO21; Archambault et al., 1996) that expresses the large subunit of RNA pol II from the repressible LEU2 promoter. The plate shown contains 2 mM leucine, threonine and isoleucine, which represses the PLEU promoter, driving low-level expression of RPB1.
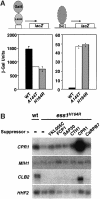
Fig. 6. Selective effects on transcription in ess1 mutant cells. (A) LacZ reporter gene expression in yeast. Left: wild-type or _ess1_ts mutant yeast were transformed with a plasmid expressing LexA–Gal4 (pSH17-4; Hanes and Brent, 1989), and a 4×-LexA operator-lacZ reporter construct (pSH18-8; S.D.Hanes and R.Brent, unpublished) at 23°C. β-galactosidase activity was measured in liquid cultures of five independent isolates. Similar results were obtained at 37°C (not shown). Right: cells were transformed with a Bicoid-site lacZ reporter (pWZ11-1; Zhu and Hanes, 2000) and treated as above, except that cells were incubated at 35°C. The differences between wild-type and the ess1 mutants were less pronounced at 23°C (not shown). (B) Northern analysis of genes encoding mitotic regulators in _ess1_H164R mutant cells. Wild-type cells or _ess1_H164R mutant cells with control vector pRS426 (–) or the indicated multicopy suppressors were grown in selective medium and shifted to 37°C for 4 h prior to harvesting. A 6 µg aliquot of total RNA was loaded per lane. Duplicate blots were hybridized with random primed 32P-labeled probes for CPR1 (cyclophilin A) and MIH1 (Cdc25 phosphatase), or CLB2 (cyclin B) and HHF2 (histone H4).
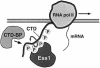
Fig. 7. Model for the action of Ess1 in transcription. Ess1 binds the phosphorylated CTD of RNA pol II, catalyzing its isomerization, thus acting as a regulatory switch for loading of proteins required for initiation, elongation, termination and 3′ end formation. In this model, Ess1 coordinates the sequential steps of transcription by changing the three-dimensional structure of the CTD, altering the affinity of protein–CTD interactions. Binding of Ess1 to the CTD would be regulated by phosphorylation–dephosphorylation by CTD kinases and CTD phosphatases (e.g. Fcp1). Ess1 might instead work stoichiometrically; Ess1 would sterically block (or promote) binding of RNA pol II-associated proteins to the phosphorylated CTD.
Similar articles
- The ESS1 prolyl isomerase and its suppressor BYE1 interact with RNA pol II to inhibit transcription elongation in Saccharomyces cerevisiae.
Wu X, Rossettini A, Hanes SD. Wu X, et al. Genetics. 2003 Dec;165(4):1687-702. doi: 10.1093/genetics/165.4.1687. Genetics. 2003. PMID: 14704159 Free PMC article. - Cyclophilin A and Ess1 interact with and regulate silencing by the Sin3-Rpd3 histone deacetylase.
Arévalo-Rodríguez M, Cardenas ME, Wu X, Hanes SD, Heitman J. Arévalo-Rodríguez M, et al. EMBO J. 2000 Jul 17;19(14):3739-49. doi: 10.1093/emboj/19.14.3739. EMBO J. 2000. PMID: 10899127 Free PMC article. - The Ess1 prolyl isomerase: traffic cop of the RNA polymerase II transcription cycle.
Hanes SD. Hanes SD. Biochim Biophys Acta. 2014;1839(4):316-33. doi: 10.1016/j.bbagrm.2014.02.001. Epub 2014 Feb 12. Biochim Biophys Acta. 2014. PMID: 24530645 Free PMC article. Review. - Functional interaction of the Ess1 prolyl isomerase with components of the RNA polymerase II initiation and termination machineries.
Krishnamurthy S, Ghazy MA, Moore C, Hampsey M. Krishnamurthy S, et al. Mol Cell Biol. 2009 Jun;29(11):2925-34. doi: 10.1128/MCB.01655-08. Epub 2009 Mar 30. Mol Cell Biol. 2009. PMID: 19332564 Free PMC article. - Prolyl isomerases in gene transcription.
Hanes SD. Hanes SD. Biochim Biophys Acta. 2015 Oct;1850(10):2017-34. doi: 10.1016/j.bbagen.2014.10.028. Epub 2014 Oct 31. Biochim Biophys Acta. 2015. PMID: 25450176 Free PMC article. Review.
Cited by
- Regulation of Skn7-dependent, oxidative stress-induced genes by the RNA polymerase II-CTD phosphatase, Fcp1, and Mediator kinase subunit, Cdk8, in yeast.
Aristizabal MJ, Dever K, Negri GL, Shen M, Hawe N, Benschop JJ, Holstege FCP, Krogan NJ, Sadowski I, Kobor MS. Aristizabal MJ, et al. J Biol Chem. 2019 Nov 1;294(44):16080-16094. doi: 10.1074/jbc.RA119.008515. Epub 2019 Sep 10. J Biol Chem. 2019. PMID: 31506296 Free PMC article. - Cleavage/polyadenylation factor IA associates with the carboxyl-terminal domain of RNA polymerase II in Saccharomyces cerevisiae.
Barillà D, Lee BA, Proudfoot NJ. Barillà D, et al. Proc Natl Acad Sci U S A. 2001 Jan 16;98(2):445-50. doi: 10.1073/pnas.98.2.445. Epub 2001 Jan 9. Proc Natl Acad Sci U S A. 2001. PMID: 11149954 Free PMC article. - Peptidyl-prolyl cis/trans isomerases and transcription: is there a twist in the tail?
Shaw PE. Shaw PE. EMBO Rep. 2007 Jan;8(1):40-5. doi: 10.1038/sj.embor.7400873. EMBO Rep. 2007. PMID: 17203101 Free PMC article. Review. - Small family with key contacts: par14 and par17 parvulin proteins, relatives of pin1, now emerge in biomedical research.
Mueller JW, Bayer P. Mueller JW, et al. Perspect Medicin Chem. 2008 Mar 7;2:11-20. doi: 10.4137/pmc.s496. Perspect Medicin Chem. 2008. PMID: 19787094 Free PMC article. - Peptidyl-prolyl isomerases: a new twist to transcription.
Shaw PE. Shaw PE. EMBO Rep. 2002 Jun;3(6):521-6. doi: 10.1093/embo-reports/kvf118. EMBO Rep. 2002. PMID: 12052773 Free PMC article. Review.
References
- Aasland R., Gibson,T.J. and Stewart,A.F. (1995) The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem. Sci., 20, 56–59. - PubMed
- Albert A., Lavoie,S. and Vincent,M. (1999) A hyperphosphorylated form of RNA polymerase II is the major interphase antigen of the phosphoprotein antibody MPM-2 and interacts with the peptidyl-prolyl isomerase Pin1. J. Cell Sci., 112, 2493–2500. - PubMed
- Archambault J., Chambers,R.S., Kobor,M.S., Ho,Y., Cartier,M., Bolotin, D., Andrews,B., Kane,C.M. and Greenblatt,J. (1997) An essential component of a C-terminal domain phosphatase that interacts with transcription factor IIF in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 94, 14300–14305. - PMC - PubMed
- Archambault J., Pan,G., Dahmus,G.K., Cartier,M., Marshall,N., Zhang,S., Dahmus,M.E. and Greenblatt,J. (1998) FCP1, the RAP74-interacting subunit of a human protein phosphatase that dephosphorylates the carboxyl-terminal domain of RNA polymerase IIO. J. Biol. Chem., 273, 27593–27601. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous