Two modes of FEN1 binding to PCNA regulated by DNA - PubMed (original) (raw)
Two modes of FEN1 binding to PCNA regulated by DNA
X V Gomes et al. EMBO J. 2000.
Abstract
The FEN1 nuclease functions during Okazaki fragment maturation in the eukaryotic cell. Like many other proliferating cell nuclear antigen (PCNA)-binding proteins, FEN1 interacts with the interdomain connector loop (IDCL) of PCNA, and PCNA greatly stimulates FEN1 activity. A yeast IDCL mutant pcna-79 (IL126,128AA) failed to interact with FEN-1, but, surprisingly, pcna-79 was still very active in stimulating FEN1 activity. In contrast, a C-terminal mutant pcna-90 (PK252,253AA) showed wild-type binding to FEN1 in solution, but poorly stimulated FEN1 activity. When PCNA was loaded onto a DNA substrate coupled to magnetic beads, it stabilized retention of FEN1 on the DNA. In this DNA-dependent binding assay, pcna-79 also stabilized retention of FEN1, but pcna-90 was inactive. Therefore, in the absence of DNA, FEN1 interacts with PCNA mainly through the IDCL. However, when PCNA encircles the DNA, the C-terminal domain of PCNA rather than its IDCL is important for binding FEN1. An FF-->GA mutation in the PCNA-interaction domain of FEN1 severely decreased both modes of interaction with PCNA and resulted in replication and repair defects in vivo.
Figures
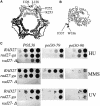
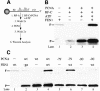
Fig. 1. Structure of PCNA and FEN1 and genetic analysis of mutants. (A) Structure of S.cerevisiae PCNA and the location of the mutations used in this study (shown in only one subunit of the homotrimer); pcna-79, IL126,128AA; and pcna-90, PK252,253AA (Krishna et al., 1994). (B) Structure of P.furiosus FEN1 and the location of the amino acids (W336 and F337) analogous to those mutated in yeast fen1-ga (FF346,347AA) (Hosfield et al., 1998). (C) Sensitivity of yeast strains to hydroxyurea and DNA-damaging agents. Serial 10-fold dilutions were made of the single or double mutants or of the isogenic wild-type yeast strain. Approximately 102, 103, 104 and 105 cells were spotted on YPDA plates containing 100 mM hydroxyurea (HU) or 0.005% MMS, or on YPDA plates and irradiated with 100 J/m2 of UV light and grown at 30°C for 3–4 days.
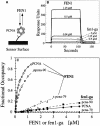
Fig. 2. Binding of FEN1 to PCNA by surface plasmon resonance. (A) Schematic of sensor chip with PCNA immobilized on the chip surface. Either wild-type PCNA, pcna-79 or pcna-90 was immobilized on the surface of a dextran B1 chip, and increasing concentrations of FEN1 or fen1-ga were passed over the chip. (B) Sensorgrams of the response of the indicated concentrations of either FEN1 or fen1-ga to immobilized PCNA. (C) Concentration–response curves for binding of FEN1 or fen1-ga to chips loaded with either PCNA (600 RU), pcna-79 (570 RU) or pcna-90 (440 RU). The data were recalculated to fractional occupancies to correct for differences in loading densities of the three PCNAs to the chip. Open symbols, FEN1 passed over the PCNA chips; filled symbols, fen1-ga passed over the chips.
Fig. 3. The C-termini of both PCNA and FEN1 are essential for FLAP cleavage at 125 mM NaCl. (A) Diagram of the DNA FLAP substrate. The position of the label is indicated by the asterisk on the FLAP strand. (B) Activity of FEN1 and fen1-ga in low salt conditions. FLAP substrate (10 fmol) was coated with 1 µg of SSB and incubated in assay buffer + 10 mM NaCl with no enzyme (lane 1), 40 or 120 fmol of FEN1 (lanes 2 and 3) or 40 or 120 fmol of fen1-ga (lanes 4 and 5). After incubation at 30°C for 3 min, the reaction products were analyzed by 15% urea–PAGE as described in Materials and methods. The mobility of the cleaved products is 19 and 20 nucleotides. (C) Stimulation of FEN1 activity by PCNA. Assays were performed in 100 µl reactions in FEN1 assay buffer + 125 mM NaCl. The FLAP substrate (10 fmol) was incubated with 1 µg of SSB, 1.4 pmol of PCNA or mutant PCNA, 2 pmol of RF-C and 100 µM ATP at 30°C for 30 s and cooled to 13°C. A 130 fmol concentration of either FEN1 or fen1-ga was added to the reaction and incubation continued at 13°C. At different time intervals, 15 µl aliquots were removed and analyzed by 15% urea–PAGE. The percentage of 5′-labeled cleavage products released was quantitated in a phosphoimager. Open symbols, assays with FEN1; filled symbols, assays with fen1-ga. The dashed line represents background activity of FEN1 in the absence of PCNA.
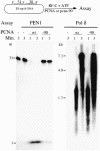
Fig. 4. Pcna-90 is active for stimulation of Pol δ but inactive for FEN1. The nick substrate as described in Materials and methods was used. The 31mer was 5′-32P-labeled. For each 30 µl assay, 10 fmol of DNA substrate were coated with 6 pmol of yeast RPA and incubated with 1 pmol of PCNA or pcna-90 as indicated, 1 pmol of RF-C and 100 µM ATP at 30°C for 30 s. Either 0.5 pmol of FEN1 and 125 mM NaCl (left panel) or 50 fmol of Pol δ together with 100 µM each of dGTP, dCTP and dTTP, 10 µM [α-32P]dATP and 75 mM NaCl (right panel) were added and 14 µl aliquots were removed after 1 and 3 min and analyzed by 15% urea–PAGE (left panel) or by 1% alkaline agarose gel electrophoresis (right panel). Controls contained no enzyme or no PCNA (lanes 1 and 2, left panel), or no PCNA (lane 1, right panel).
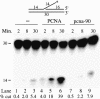
Fig. 5. pcna-90 does not stimulate FEN1 activity on an oligonucleotide FLAP. The substrate used in the assay has been described (Li et al., 1995). The assay was performed in a 50 µl reaction containing 50 mM Tris–HCl pH 8.1, 10 mM MgCl2, 1 mM DTT, 0.5 mg/ml BSA, 25 mM NaCl, 50 fmol of FLAP substrate, 100 fmol of FEN1 and either no PCNA, 5.7 pmol of PCNA or 5.7 pmol of pcna-90 as indicated. After various times at 30°C, 15 µl aliquots were analyzed by 15% urea–PAGE. The percentage of FLAP cut was quantitated and is given below each lane.
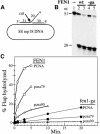
Fig. 6. pcna-79, but not pcna-90 stabilizes binding of FEN1 on DNA. (A) Outline of the assay. The DNA was attached to magnetic beads via a biotin–streptavidin linkage. The nick substrate was incubated in a stepwise fashion with the indicated proteins and processed for immunoblot analysis. (B) Binding of PCNA requires ATP (lanes 2 and 3) and binding of FEN1 requires PCNA (lanes 1 and 4). The positions of FEN1 (F) and PCNA (P) are indicated. (C) Stable complexes are not formed with fen1-ga (lane 2), nor with pcna-90 (lane 8). For details see Materials and methods and Results.
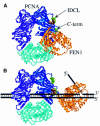
Fig. 7. A two-domain model for interaction between PCNA and FEN1. (A) A Rasmol ribbon diagram of yeast PCNA is shown with the three identical subunits indicated in blue, purple and cyan. Ile126 and Leu128 in the IDCL (mutated to Ala in pcna-79), and Pro252 and Lys253 in the C-terminus (mutated to Ala in pcna-90) are shown as space-filling residues in green. The donut structure is rotated 45° out of the plane of the paper. The P.furiosus FEN1 in gold with the Phe–Trp dipeptide of the PCNA-interacting domain as a space-filling model in brown was modeled into the PCNA structure analogously to the human PCNA–p21 co-crystal structure (Gulbis et al., 1996). (B) With PCNA encircling the DNA, binding of FEN1 shifts to the C-terminus of PCNA. A model of the interaction on a FLAP substrate is shown.
Similar articles
- Homologous regions of Fen1 and p21Cip1 compete for binding to the same site on PCNA: a potential mechanism to co-ordinate DNA replication and repair.
Warbrick E, Lane DP, Glover DM, Cox LS. Warbrick E, et al. Oncogene. 1997 May 15;14(19):2313-21. doi: 10.1038/sj.onc.1201072. Oncogene. 1997. PMID: 9178907 - Regulation of DNA replication and repair proteins through interaction with the front side of proliferating cell nuclear antigen.
Jónsson ZO, Hindges R, Hübscher U. Jónsson ZO, et al. EMBO J. 1998 Apr 15;17(8):2412-25. doi: 10.1093/emboj/17.8.2412. EMBO J. 1998. PMID: 9545252 Free PMC article. - p21Cip1/Waf1 disrupts the recruitment of human Fen1 by proliferating-cell nuclear antigen into the DNA replication complex.
Chen U, Chen S, Saha P, Dutta A. Chen U, et al. Proc Natl Acad Sci U S A. 1996 Oct 15;93(21):11597-602. doi: 10.1073/pnas.93.21.11597. Proc Natl Acad Sci U S A. 1996. PMID: 8876181 Free PMC article. - PCNA binding through a conserved motif.
Warbrick E. Warbrick E. Bioessays. 1998 Mar;20(3):195-9. doi: 10.1002/(SICI)1521-1878(199803)20:3<195::AID-BIES2>3.0.CO;2-R. Bioessays. 1998. PMID: 9631646 Review. - Structural and Functional Insight into Proliferating Cell Nuclear Antigen.
Park SY, Jeong MS, Han CW, Yu HS, Jang SB. Park SY, et al. J Microbiol Biotechnol. 2016 Apr 28;26(4):637-47. doi: 10.4014/jmb.1509.09051. J Microbiol Biotechnol. 2016. PMID: 26699741 Review.
Cited by
- Disruption of the FEN-1/PCNA interaction results in DNA replication defects, pulmonary hypoplasia, pancytopenia, and newborn lethality in mice.
Zheng L, Dai H, Qiu J, Huang Q, Shen B. Zheng L, et al. Mol Cell Biol. 2007 Apr;27(8):3176-86. doi: 10.1128/MCB.01652-06. Epub 2007 Feb 5. Mol Cell Biol. 2007. PMID: 17283043 Free PMC article. - Histone deposition protein Asf1 maintains DNA replisome integrity and interacts with replication factor C.
Franco AA, Lam WM, Burgers PM, Kaufman PD. Franco AA, et al. Genes Dev. 2005 Jun 1;19(11):1365-75. doi: 10.1101/gad.1305005. Epub 2005 May 18. Genes Dev. 2005. PMID: 15901673 Free PMC article. - Early immobilization of nuclease FEN1 and accumulation of hRAD18 protein at stalled DNA replication forks in mammalian cells.
Nikiforov AA, Sasina LK, Svetlova MP, Solovjeva LV, Oei SL, Bradbury EM, Tomilin NV. Nikiforov AA, et al. Dokl Biochem Biophys. 2003 Mar-Apr;389:122-5. doi: 10.1023/a:1023696425171. Dokl Biochem Biophys. 2003. PMID: 12856420 No abstract available. - Structural and functional studies of PCNA from African swine fever virus.
Shao Z, Yang J, Gao Y, Zhang Y, Zhao X, Shao Q, Zhang W, Cao C, Liu H, Gan J. Shao Z, et al. J Virol. 2023 Aug 31;97(8):e0074823. doi: 10.1128/jvi.00748-23. Epub 2023 Aug 3. J Virol. 2023. PMID: 37534905 Free PMC article. - A novel mechanism for regulating the activity of proliferating cell nuclear antigen by a small protein.
Li Z, Huang RY, Yopp DC, Hileman TH, Santangelo TJ, Hurwitz J, Hudgens JW, Kelman Z. Li Z, et al. Nucleic Acids Res. 2014 May;42(9):5776-89. doi: 10.1093/nar/gku239. Epub 2014 Apr 11. Nucleic Acids Res. 2014. PMID: 24728986 Free PMC article.
References
- Bambara R.A., Murante,R.S. and Henricksen,L.A. (1997) Enzymes and reactions at the eukaryotic DNA replication fork. J. Biol. Chem., 272, 4647–4650. - PubMed
- Burgers P.M. and Gerik,K.J. (1998) Structure and processivity of two forms of Saccharomyces cerevisiae DNA polymerase δ. J. Biol. Chem., 273, 19756–19762. - PubMed
- Burgers P.M.J. and Yoder,B.L. (1993) ATP-independent loading of the proliferating cell nuclear antigen requires DNA ends. J. Biol. Chem., 268, 19923–19936. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous